OPTIMIZATION OF FISH ENVIRONMENTAL DNA SAMPLE PROCESSING AND PRESERVATION TECHNOLOGY BASED ON DROPLET DIGITAL PCR
-
摘要: 以实验室内的鲫(Carassius auratus)为研究对象, 利用微滴式数字PCR(Droplet Digital PCR, ddPCR)定量技术, 优化了鱼类环境DNA(Environmental DNA, eDNA)样本的捕获、提取和保存方法, 并对免DNA提取的 PCR直扩技术进行了探索。研究结果如下: (1)在同一孔径、不同材质的6种滤膜中, 捕获总量最高的是混合纤维素膜, 最低的是聚碳酸酯膜, 前者获得的ddPCR产物浓度可达后者的17倍; (2)提取DNA总量最高的是Qiagen DNeasy PowerWater kit, 最低的是高盐法, 前者获得的ddPCR产物浓度是后者的5.5倍; (3)不同滤膜和提取方法对最终的ddPCR产物总量有显著的交互作用(P<0.001); 混合纤维素膜过滤和Qiagen DNeasy PowerWater kit提取的组合效果显著优于其他组合; (4)滤膜在–20℃冷冻保存和利用Longmire’s buffer保存液保存的效果最好; (5)免DNA提取PCR直扩实验结果表明, 高保真酶可以直接对水样进行扩增。研究对eDNA操作流程中的关键步骤进行了详细比较, 确定了鱼类eDNA样本处理与保存的最优方案, 为建立eDNA标准化实验流程提供了参考依据。Abstract: In this study, the methods of fish environmental DNA (eDNA) capture, extraction and preservation were optimized by Droplet Digital PCR (ddPCR) quantitative technology using Carassius auratus in the laboratory, and the direct PCR technology without DNA extraction was preliminarily explored. The results showed that the mixed cellulose ester membrane produced the most ddPCR product, among the six kinds of filter membrane, and the polycarbonate membrane produced the lowest ddPCR product with only 1/17 of mixed cellulose ester membrane. The method which extracted the highest amount of DNA was the Qiagen DNeasy PowerWater kit, and the lowest was the high-salt method. The Qiagen DNeasy PowerWater kit yielded better results compared to the high-salt method, showing a 5.5-fold improvement in DNA yield. Different filter membranes and extraction methods have significant interactions on the total amount of final ddPCR product (P<0.001), filtration through the mixed cellulose ester membranes and Qiagen DNeasy PowerWater kit significantly outperformed other combinations of capture and extraction methods. Filter membranes stored at –20℃ or stored in Longmire's buffer solution recovered the most ddPCR product. The results of direct PCR amplification without DNA extraction showed that the high-fidelity enzyme of Vazyme can directly amplify the water samples and obtain the target DNA product. This study, compared the key steps in the eDNA operation process in detail, and the optimal protocol of fish eDNA sample processing and preservation was determined, which provided a reference for the establishment of eDNA standardized experimental process.
-
Keywords:
- Environmental DNA /
- Filter /
- Extraction method /
- Preservation condition
-
全球生物多样性下降是21世纪面临的最为严峻的挑战之一[1, 2]。在此背景下, 开展全面准确的生物多样性监测对于生物多样性保护具有重要意义。传统的生物多样性监测主要基于实地考察等方法, 需要消耗大量的人力、物力和财力, 同时也会对研究对象和采样生境造成一定的破坏[3, 4], 且依赖于具有分类学专业知识的研究人员。
近年来, 环境DNA(environmental DNA, eDNA)技术作为一种新型、无创、经济高效的监测方法受到广泛关注[5], 可替代或辅助传统监测方法, 大大提高生物多样性调查和研究的效率[6, 7]。eDNA是指从环境样品(如水、土壤、沉积物等)中提取到的DNA片段总和, 既包含生物经由皮肤、尿液、粪便和黏液等释放到环境中表皮细胞中的胞内DNA, 也包括细胞死亡后裂解释放到环境中的胞外DNA [8, 9]。通过对这些eDNA片段或是基因组进行PCR扩增和高通量测序, 并根据已有的DNA数据库进行比对注释, 可以实现对所有生物群落的监测。目前, 该技术在水体生物多样性调查等方面已开展广泛应用, 例如: (1)检测目标物种的有无。在入侵物种、濒危物种及其他稀有物种的检测中具有广泛应用, 如监测亚洲鲤(Asian carps)[10]的入侵, 中华鲟(Acipenser sinensis)的洄游路线[11]; (2)进行生物量的估测。研究表明, 水体中eDNA 的量与生物量存在正相关, 可以推断水体中生物的种群密度[12, 13]; (3)生物多样性评估[8, 14, 15]等。例如, 利用eDNA宏条形码评估河口鱼类的多样性[16]。
eDNA实验过程一般包括野外样品采集、eDNA捕获和保存、总DNA提取3个步骤[17]。选择不同的eDNA捕获和提取方法会显著影响样品中获得的DNA浓度或产量[18—21], 并进一步影响生物量估算和生物多样性分析的结果[22]。在水生态系统研究中, eDNA的捕获通常采用过滤法[18—21]。例如, Eichmiller等[20]对eDNA捕获方法中的沉淀法、离心法和过滤法进行了研究, 结果表明过滤方法得到的eDNA产量最多。但针对不同的样品来源及不同的研究类群, eDNA的捕获及实验方法各具特点, 缺乏统一的标准, 降低了不同研究之间的可比性。例如: Renshaw等[23]通过CTAB buffer保存和酚氯仿异戊醇提取法对四种滤膜材料(硝酸纤维素膜、聚醚砜膜、聚碳酸酯膜、玻璃纤维素膜)捕获eDNA效果的研究结果表明, 使用同一孔径的硝酸纤维素膜和聚醚砜膜的捕获结果最优, 且基于这两种滤膜获得的DNA产量不存在显著差异。 但是Hinlo等[18]通过–20℃冷冻保存和Qiagen DNeasy Blood & Tissue kit提取方法对4种滤膜材料(玻璃纤维素膜、硝酸纤维素膜、聚醚砜膜和聚碳酸酯膜)捕获eDNA效果的研究表明, 硝酸纤维素膜的捕获结果最优, 与聚醚砜膜过滤获得的DNA产量在统计学上存在显著差异。这两项研究的结果的差异可能就是由于使用的DNA提取方法或保存方法的不同组合造成的。此外, 他们的研究中没有对混合纤维素膜的捕获效果进行比较。Liang等[24]通过4℃冷藏和PowerSoil DNA kit(MoBio公司)提取方法对4种滤膜材料(聚偏氟乙烯膜、聚碳酸酯膜、聚醚砜膜、混合纤维素膜)捕获eDNA效果的研究表明, 混合纤维素膜的捕获效果最优。但是Liang等[24]并没有对硝酸纤维素膜的捕获效果进行比较。综上所述, 已有的一些研究涉及到的滤膜种类和DNA提取方法不多, 不足以提供全面的参考。因此, 为了提高eDNA技术在生物多样性监测中的应用, 亟需对eDNA捕获、保存和提取方法进行优化和标准化。
基于定量PCR, eDNA技术不仅可以检测水样中目标物种是否存在, 还可以通过量化eDNA对其物种丰度和生物量等进行估算。微滴式数字PCR(Droplet Digital PCR, ddPCR)技术, 是在实时荧光定量PCR(Real-time fluorescence quantitative PCR, qPCR)技术基础上发展起来的一种新的核酸定性定量检测方法, 可以直接检测样品中的目标基因的绝对拷贝数, 称为“第3代PCR”技术[25]。基于qPCR的eDNA分析对抑制剂比较敏感[26—28], 容易造成假阴性。ddPCR不依赖于扩增曲线的循环阈值进行定量, 不受扩增效率影响, 同时更适合检测微量DNA, 对PCR抑制剂耐受性好, 不需要依赖参考曲线 [29], 具有比qPCR更高的灵敏度和精确度[26, 30]。
本研究以实验室内的鲫(Carassius auratus)为对象, 对获取水样eDNA的滤膜材料和DNA提取方案进行组合测试, 并利用ddPCR定量技术比较不同组合的效率; 同时对滤膜保存方法和免DNA提取的PCR直扩实验进行了初步探索, 优化鱼类eDNA样本处理与保存方案, 从而促进eDNA技术在水生生物物种多样性监测和保护中的应用。
1. 材料与方法
1.1 实验室内环境DNA样品的采集
2020年8月于武汉市武昌区花鸟市场购买80尾体格健壮、规格一致的鲫。平均体长为 (61.25±4.56) mm, 体重为(8.19±1.71) g。首先, 用10% 漂白粉溶液对145 cm×70 cm×60 cm的水族缸进行清洗消毒。随后注入304 L容量的水, 将80尾鲫放入缸中, 所有鱼类在放入水族缸前暂养3d, 不进行投喂, 避免受到其他物种eDNA污染。实验用水是经24h曝气后的自来水, 并用充气泵不断向水缸中泵入气体。同时设置一同体积的空白水缸(只放同体积的水不放鱼), 在实验时取空白水缸的水样作为空白对照来评估水缸是否受到外源污染。
在进行水样采集之前, 用超纯水将500 mL的采样瓶、采水器先润洗两遍, 再用10%漂白粉溶液浸泡10min, 最后用超纯水润洗两遍。80尾鲫在水族缸暂养3d后, 将水族缸内水混匀后采集水样。清洗、采水时应佩戴一次性手套。水样采集后立即置于4℃保存。
1.2 不同材质滤膜DNA捕获效率的比较
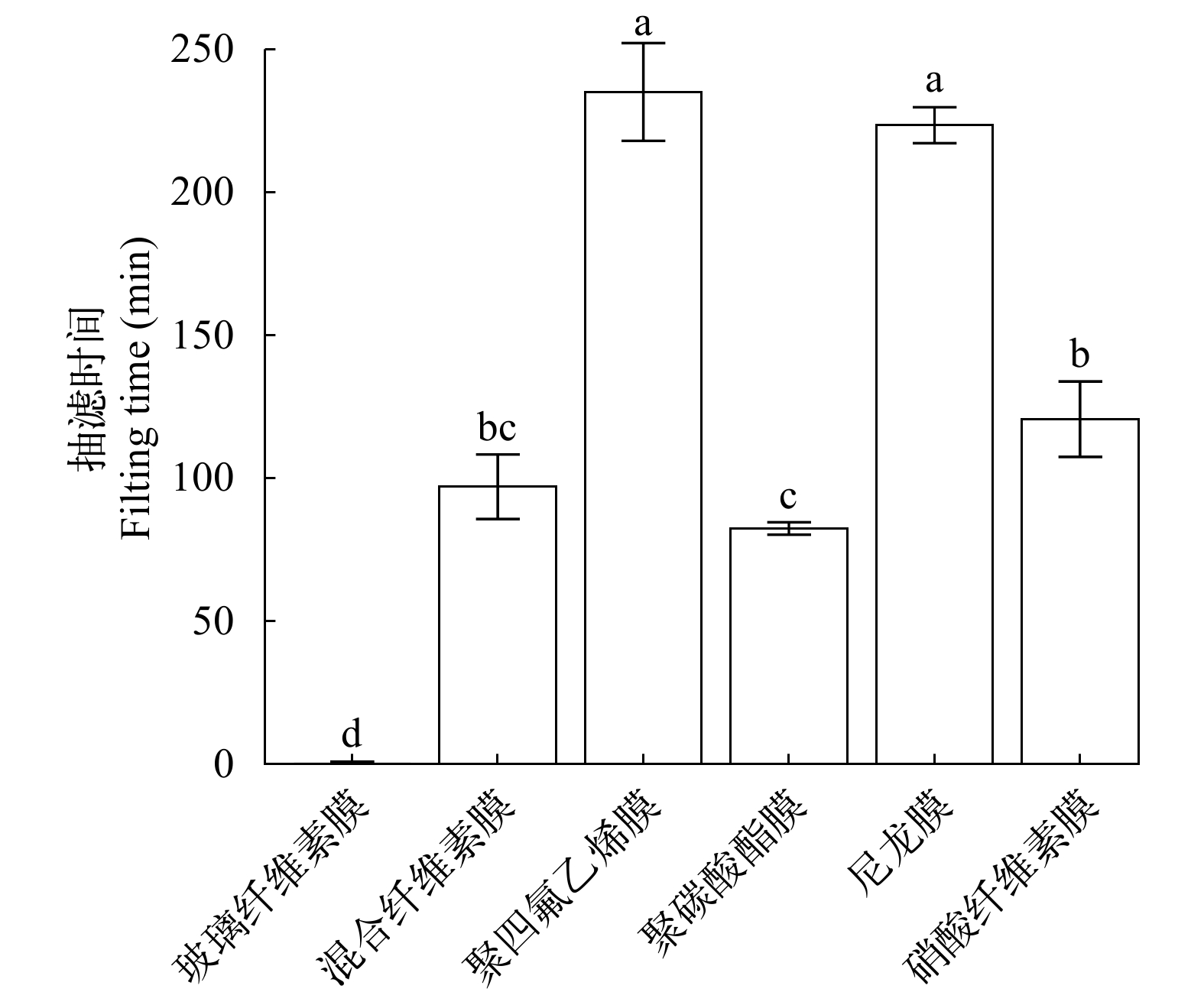
比较常见的6种不同材质滤膜的DNA捕获总量(硝酸纤维素膜-Cellulose-nitrate、玻璃纤维素膜-Glass-fiber、聚碳酸酯膜-Polycarbonate、聚四氟乙烯膜-Poly tetra fluoroethylene、混合纤维素膜-Mixed cellulose ester 和尼龙膜-Nylon), 这些滤膜均产自上海市新亚净化器件厂, 滤膜孔径大小均为0.22 μm, 直径为50 mm。每种滤膜各抽滤3个重复, 并取曝气的自来水和超纯水作为空白对照, 每个重复采集500 mL放有鲫的水样, 总计18份水样和12份空白对照。使用津腾-隔膜真空泵(型号: GM-0.33A)进行抽滤, 同时记录抽滤时间, 抽滤结束后, 使用灭菌的镊子从滤膜的边缘夹起, 将滤膜卷成长条状放入50 mL已灭菌的离心管内, 置于–20℃保存直至eDNA提取。eDNA提取统一采用Qiagen DNeasy PowerWater Kit, 确保该实验中只有eDNA捕获方法有差异, 从而对滤膜的捕获性能进行比较, 筛选出最佳捕获效率的滤膜材料。
1.3 不同DNA提取方法的比较
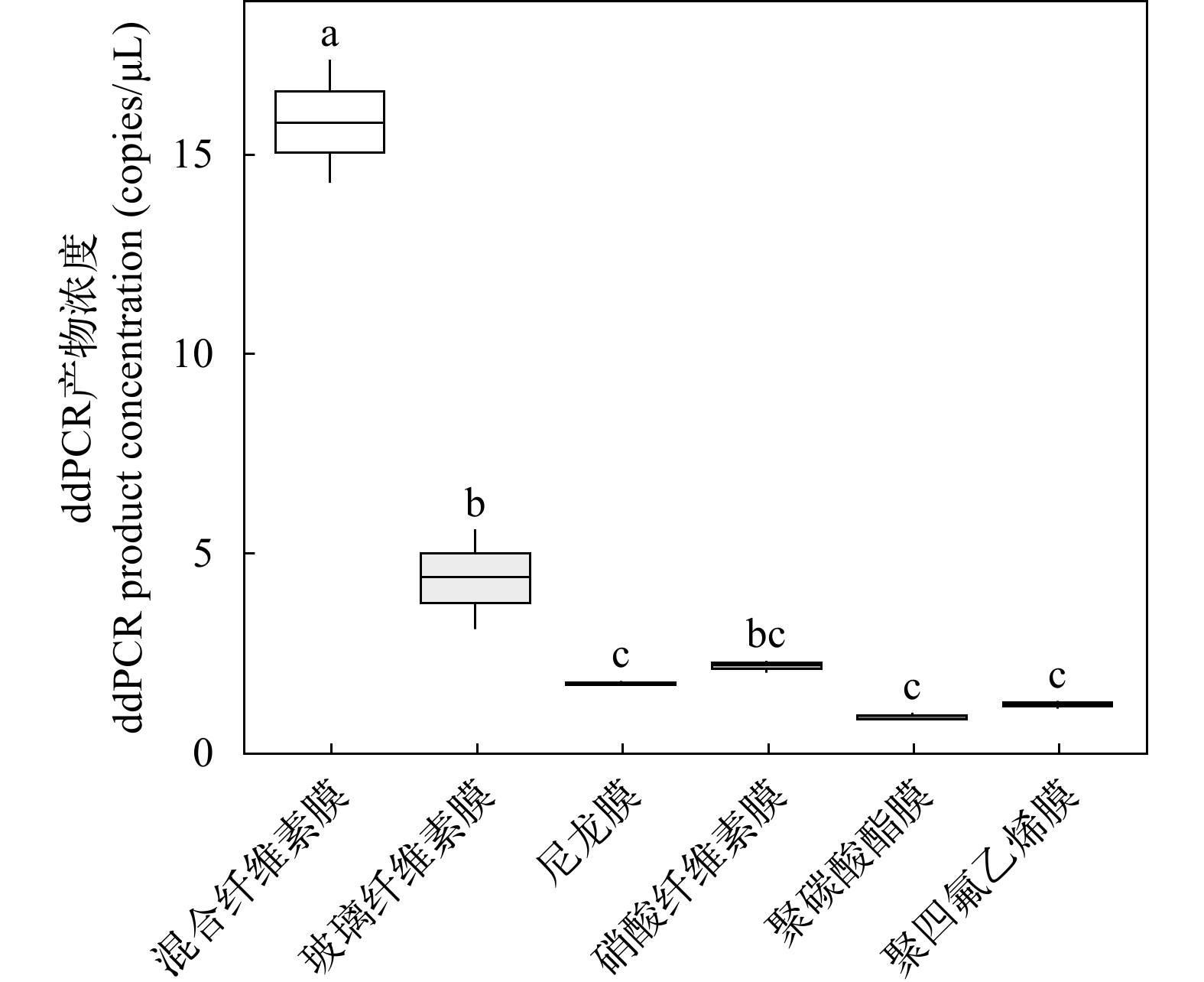
用5种不同的DNA提取方法进行实验(酚氯仿异戊醇法、高盐法、Qiagen DNeasy Blood & Tissue Kit、磁珠法和Qiagen DNeasy PowerWater Kit): (1)改良的酚氯仿异戊醇法, 使用乙醇进行沉淀[23, 31, 32]。(2)高盐法, 参照Aljanabi和Martinez[33]并对原方法进行了3处改动: ①将滤膜置于2 mL离心管内剪碎, 加 500 μL HOM Buffer 和 20 μL 蛋白酶 K, 于55℃消化3h(每隔1h涡旋10min); ②加入500 μL NaCl(4.5 mol/L), 300 μL氯仿, 混匀15min, 10000 r/min离心10min, 将全部溶液转移至新离心管内, 弃去滤膜; ③将上述溶液以10000 r/min离心10min, 抽取上清液。(3)Qiagen DNeasy Blood & Tissue Kit法, 参照Qiagen DNeasy Blood & Tissue Kit[23]进行实验操作。(4) 磁珠法, 参照Magnetic Animal Tissue Genomic DNA Kit(天根)说明书并稍做改动: ①将滤膜剪碎置于2 mL离心管内, 加入400 μL GHA和30 μL蛋白酶K于55℃消化3h(每隔1h涡旋10min), 消化结束后12000 r/min离心5min, 取出全部溶液置于新离心管内, 弃滤膜; ②加入400 μL GHB, 振荡混匀。(5)Qiagen DNeasy PowerWater Kit, 参照试剂盒说明书操作。根据eDNA捕获实验的结果, 使用DNA捕获总量最高的混合纤维素膜(孔径0.22 μm, 直径50 mm)对水样进行抽滤。每种eDNA提取方法各抽取3个重复, 曝气的自来水和超纯水作空白对照, 共计15份水样和10份空白对照。最终用100 μL ddH2O洗脱DNA, 提取的eDNA置于–20℃保存备用, 直至下一步分析。
1.4 不同材质滤膜和DNA提取方法的交叉组合比较
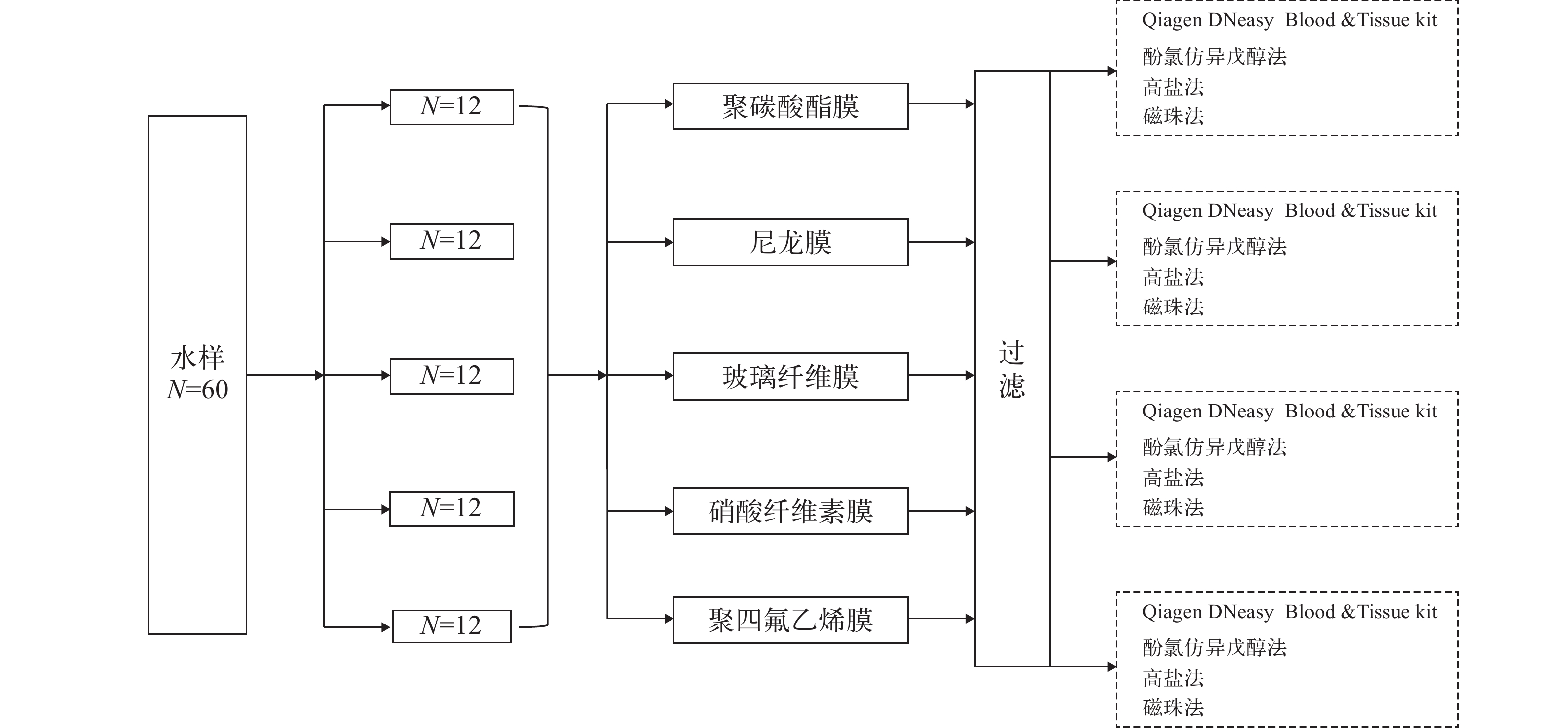
为全面比较不同滤膜类型和DNA提取方法对ddPCR产物浓度的影响, 进一步对不同材料滤膜和DNA提取方法进行交叉组合实验(图 1)。另取试验水缸的水样60份(500 mL/份), 采用余下的相同孔径的五种材质滤膜(硝酸纤维素膜、玻璃纤维素膜、聚碳酸酯膜、聚四氟乙烯膜和尼龙膜)对水样进行抽滤。每种滤膜各抽滤12个水样和8个空白对照, 分别用其余4种DNA提取方法(酚氯仿异戊醇法、高盐法、磁珠法、Qiagen DNeasy Blood & Tissue Kit)提取eDNA。
1.5 滤膜不同保存方法的比较
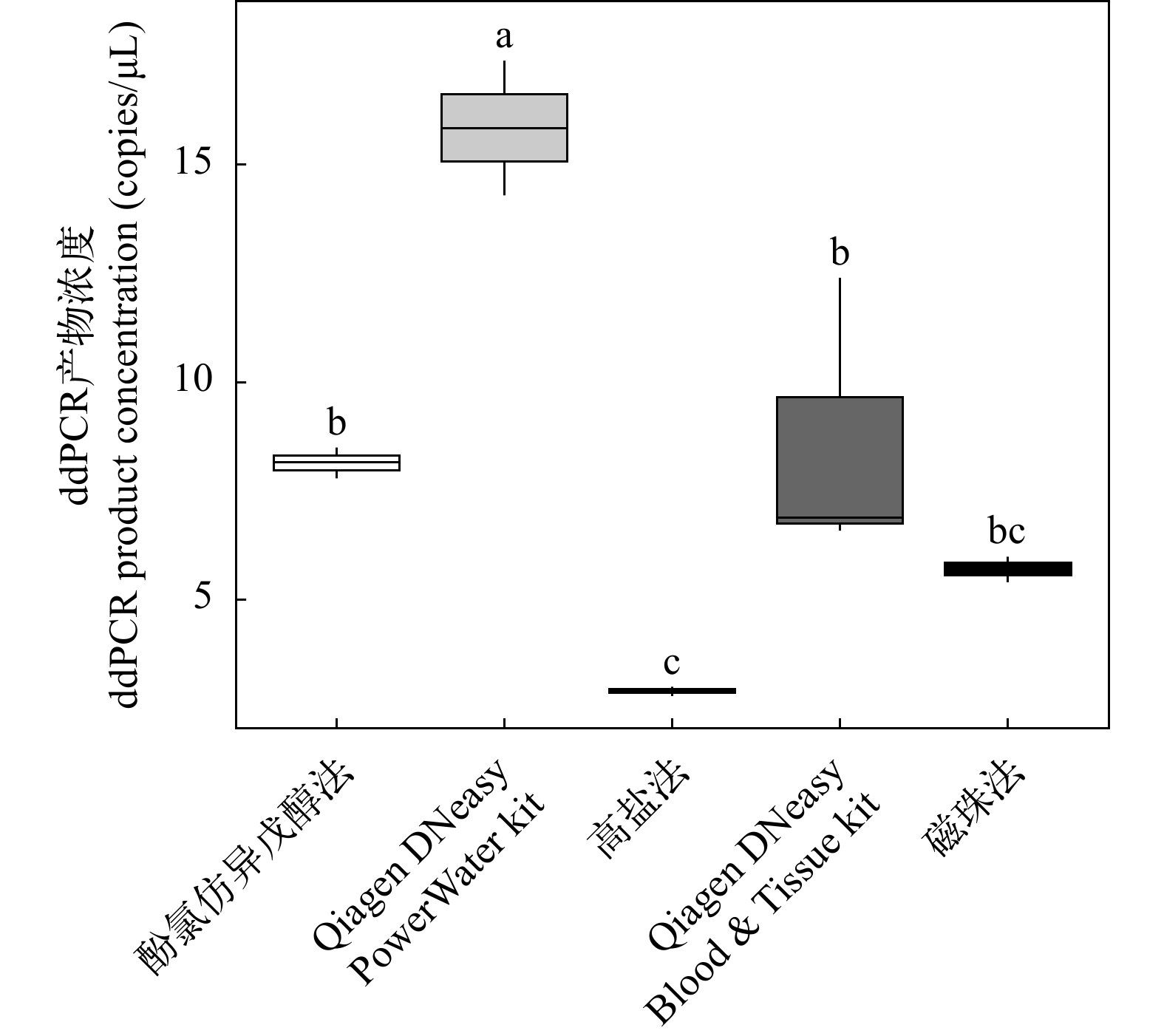
基于试验水缸的水样, 使用混合纤维素膜抽滤, Qiagen DNeasy PowerWater kit进行DNA提取。对常用的滤膜保存方法进行比较, 包括–20℃冷冻、室温干燥、RNAlater保存液、CTAB buffer[34]、Longmire’s buffer[35]及不同浓度乙醇(75%乙醇、85%乙醇、95%乙醇和无水乙醇), 其中后8种保存方法均为室温条件保存。每种保存方法抽滤3个重复和一个超纯水空白对照, 共需54份水样和18份空白对照, 将滤膜放置于不同保存条件下24h和72h后分别进行DNA提取。
1.6 免DNA提取PCR直扩研究
以试验水缸的水样为研究对象, 部分样本辅以酒精沉淀法进行预处理[36—38], 实验步骤为: (1)15 mL水样+1.5 mL 3 mol/L醋酸钠(pH=5.2)+30 mL乙醇, 4℃下10000×g离心1h去掉上清液; (2)用100 μL超纯水重悬沉淀2次, 最后保留200 μL的溶液作为PCR直扩的DNA模板。采用直接添加不同体积的无处理水样(5—21 μL)、酒精沉淀预处理水样(5—21 μL)当模板, 对TaKaRa免抽提试剂盒(TerraTM PCR Direct Polymerase Mix)、诺唯赞高保真酶(P505-d1)、诺唯赞高效酶(P213-AA)和生工直接PCR扩增试剂盒4种免DNA提取PCR直扩方法进行比较, 找出最佳的PCR反应体系。最后, 利用武汉市武昌区东湖的水样来验证上述实验室水样免DNA提取PCR直扩方法的有效性。
1.7 DNA产量测定和数据分析
本实验使用ddPCR和Bylemans等[39] 针对线粒体12S rRNA基因开发设计的鱼类特异性引物“AcMDB07”(AcMDB07-F:5′-GCCTATATACCGCCGTCG-3′; AcMDB07-R: 5′-GTACACTTACCATGTTACGACTT-3′)对每个样本中提取的鲫eDNA浓度进行定量, 扩增产物片段为321 bp。样品eDNA的检测与定量估算在中国科学院水生生物研究所分析测试中心的Bio-Rad QX200TM AutoDGTM Droplet DigitalTM PCR系统上进行。 20 μL的ddPCR扩增体系为: 2× ddPCR Super Mix for SYBR Green 为10 μL, AcMDB07-F和AcMDB07-R各为2 μL, DNA模板为2 μL, 无菌双蒸水为4 μL。PCR过程的反应程序为: 95℃, 10min; 95℃ 10s, 55℃ 1min, 共40个循环; 4℃, 5min; 90℃, 5min; 4℃保存反应产物。每个样品进行3次ddPCR重复, 每板设置阳性对照和阴性对照。在PCR扩增结束后, 将产物平稳地放入微滴读取仪中, 微滴读取仪器依次对微滴进行荧光检测, 并对每一个微滴的相对荧光值进行记录, 程序自动计算出每一反应产物中的模板浓度(copies/μL), 每个样品的ddPCR产物浓度为3次重复ddPCR扩增的平均浓度。在ddPCR定量检测后, 随机抽取了9个样本进行PCR测序, 测序结果与GenBank数据库比对发现是鲫的DNA片段。
所有数据统计分析均在R软件中进行, 对不同处理间的捕获、提取和保存方法的效果进行单因素方差分析(One-way ANOVA), 对捕获和DNA提取交叉组合实验进行双因素方差分析(Two-way ANOVA), 对主效应方差分析和交互效应方差分析模型进行对比, 佐证交互效应是否显著, 从而筛选出鱼类eDNA样本处理的最优方案。
2. 结果
2.1 不同材质滤膜对eDNA提取的影响
随机抽取的PCR检测结果表明ddPCR产物均为鲫的目标片段。通过对6种不同材质滤膜的水样抽滤时间和ddPCR产物浓度进行比较, 结果发现, 玻璃纤维素膜的抽滤时间最短(<1min), 其次为聚碳酸酯膜、混合纤维素膜、硝酸纤维素膜, 尼龙膜和聚四氟乙烯膜的抽滤时间最长(图 2)。不同材质滤膜对总ddPCR产物浓度有显著的影响(F=148.40, P<0.001; 图 3)。混合纤维素膜捕获效果显著高于其他五种材质滤膜(P<0.001), ddPCR产物浓度为(15.85±2.19) copies/μL; 玻璃纤维素膜捕获效果仅次于混合纤维素膜, 为(4.37±1.25) copies/μL(P<0.001); 硝酸纤维膜、尼龙膜和聚四氟乙烯膜3种滤膜抽滤的ddPCR产物浓度相当(1.21—2.17 copies/μL, P>0.05), 抽滤效果最差的是聚碳酸酯膜, 总ddPCR产物浓度为(0.89±0.10) copies/μL。混合纤维素膜捕获的ddPCR产物浓度是聚碳酸酯膜捕获的17倍。综合来看, 混合纤维素膜的ddPCR产物浓度最高, 但抽滤时间较长; 玻璃纤维素膜抽滤时间最短, 但是ddPCR产物浓度并没有混合纤维素膜高。
2.2 DNA不同提取方法的比较
通过随机抽取扩增后产物进行检测, 结果表明ddPCR产物均为鲫的目标片段。单因素方差分析显示(图 4), DNA不同提取方法对总ddPCR产物浓度有显著的影响(F=26.26, P<0.001)。Qiagen DNeasy PowerWater kit提取的ddPCR产物浓度最高, 平均为(15.85±2.19) copies/μL, 显著高于Qiagen DNeasy Tissue & Blood kit [(8.63±3.27) copies/μL](P<0.002)和酚氯仿异戊醇法[(8.15 ± 0.50) copies/μL](P<0.002)。高盐法提取的ddPCR产物浓度最低, 平均为(2.89±0.59) copies/μL。Qiagen DNeasy PowerWater kit提取的ddPCR产物浓度是高盐法提取的5.5倍。
2.3 不同材质滤膜和DNA提取方法组合比较
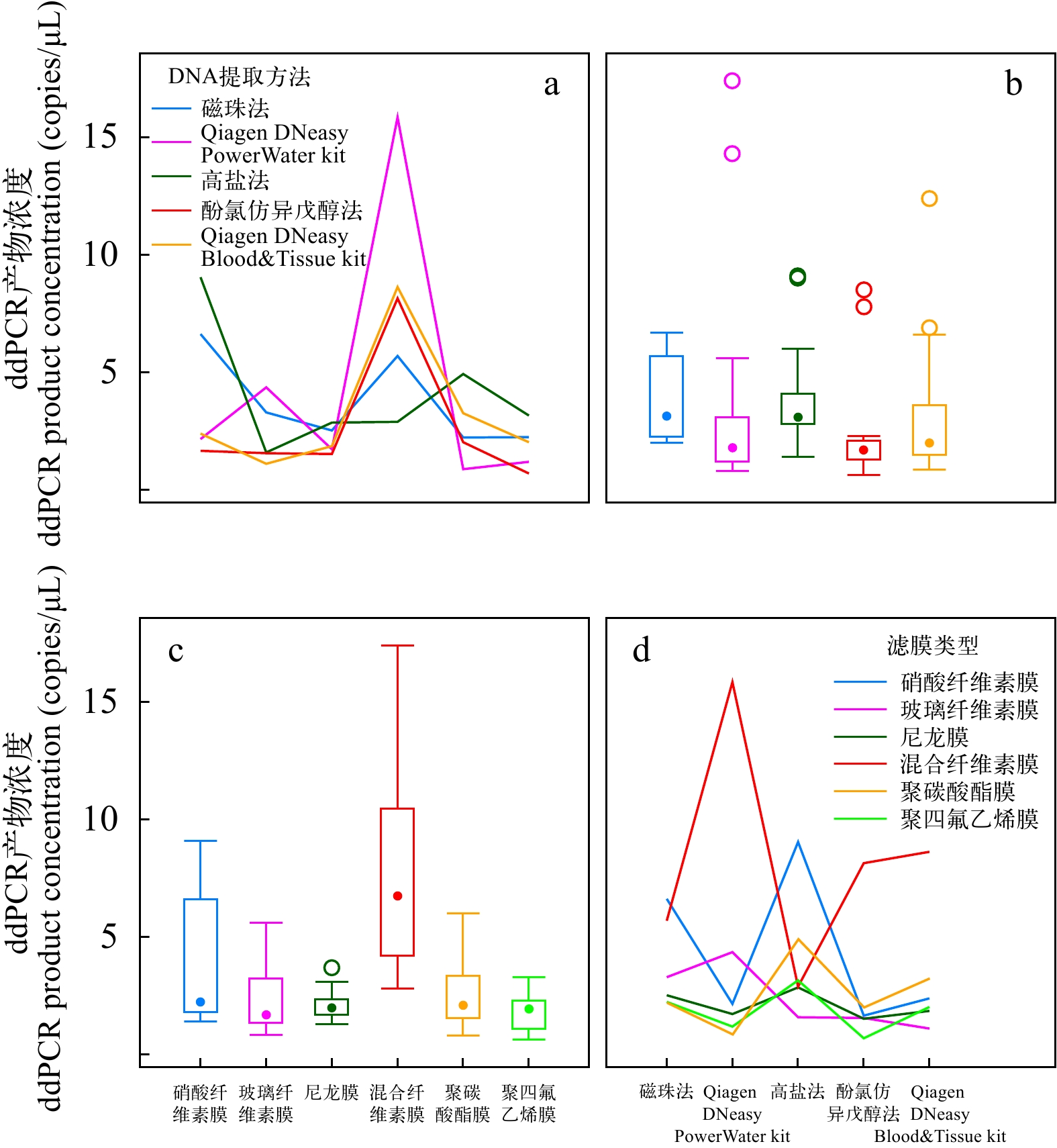
本实验对同一孔径的6种不同材质滤膜和5种不同eDNA提取方法进行交叉组合实验, 双因素方差分析及交互效应分析结果表明不同捕获和提取方法对最终的ddPCR产物浓度有显著的交互作用(F=20.59, P<0.001; 图 5)。主效应分析结果显示, 在不考虑提取方法因素时, 滤膜的ddPCR产物浓度为: 混合纤维素膜>硝酸纤维素膜>聚碳酸酯膜>玻璃纤维素膜>尼龙膜>聚四氟乙烯膜(图 5c); 在不考虑滤膜因素时, DNA提取方法的ddPCR产物浓度为: 磁珠法>高盐法>Qiagen DNeasy Blood & Tissue kit>Qiagen DNeasy PowerWater kit >酚氯仿异戊醇法(图 5b), 但在考虑到6种不同滤膜和DNA提取方法的交互作用后, DNA提取方法的ddPCR产物浓度为Qiagen DNeasy PowerWater kit>高盐法> Qiagen DNeasy Blood & Tissue kit>磁珠法>酚氯仿异戊醇法(图 5d)。综上所述, 混合纤维素膜和Qiagen DNeasy PowerWater kit组合获得的ddPCR产物浓度最高, 为(15.85±2.19) copies/μL; 聚四氟乙烯膜和酚氯仿异戊醇法组合的ddPCR产物浓度最低, 为(0.70±0.07) copies/μL, 最高组合浓度是最低浓度组合的22倍。
![]() 图 5 滤膜和eDNA提取方法组合比较a. 同一滤膜类型在不同DNA提取方法下获得的ddPCR产物浓度; b. DNA提取方法的主效应图, 即不考虑滤膜, 所有样品用不同DNA提取方法获得的ddPCR产物浓度; c. 滤膜类型的主效应图, 即不考虑提取方法, 所有样品用不同类型滤膜过滤获得的ddPCR产物浓度; d. 同一提取方法在不同滤膜过滤情况下获得的ddPCR产物浓度Figure 5. The comparison of cross-combination of filters and DNA extraction methodsa. The ddPCR concentration obtained by the same filter under different DNA extraction methods; b. The main effect of the DNA extraction methods. The ddPCR concentration obtained by different DNA extraction methods for all samples without considering the filters; c. The main effect of the filter types, The ddPCR concentration obtained by different filter types for all samples without considering the DNA extraction methods; d. The ddPCR concentration obtained by the same extraction method under different filters
图 5 滤膜和eDNA提取方法组合比较a. 同一滤膜类型在不同DNA提取方法下获得的ddPCR产物浓度; b. DNA提取方法的主效应图, 即不考虑滤膜, 所有样品用不同DNA提取方法获得的ddPCR产物浓度; c. 滤膜类型的主效应图, 即不考虑提取方法, 所有样品用不同类型滤膜过滤获得的ddPCR产物浓度; d. 同一提取方法在不同滤膜过滤情况下获得的ddPCR产物浓度Figure 5. The comparison of cross-combination of filters and DNA extraction methodsa. The ddPCR concentration obtained by the same filter under different DNA extraction methods; b. The main effect of the DNA extraction methods. The ddPCR concentration obtained by different DNA extraction methods for all samples without considering the filters; c. The main effect of the filter types, The ddPCR concentration obtained by different filter types for all samples without considering the DNA extraction methods; d. The ddPCR concentration obtained by the same extraction method under different filters2.4 滤膜保存方法的比较
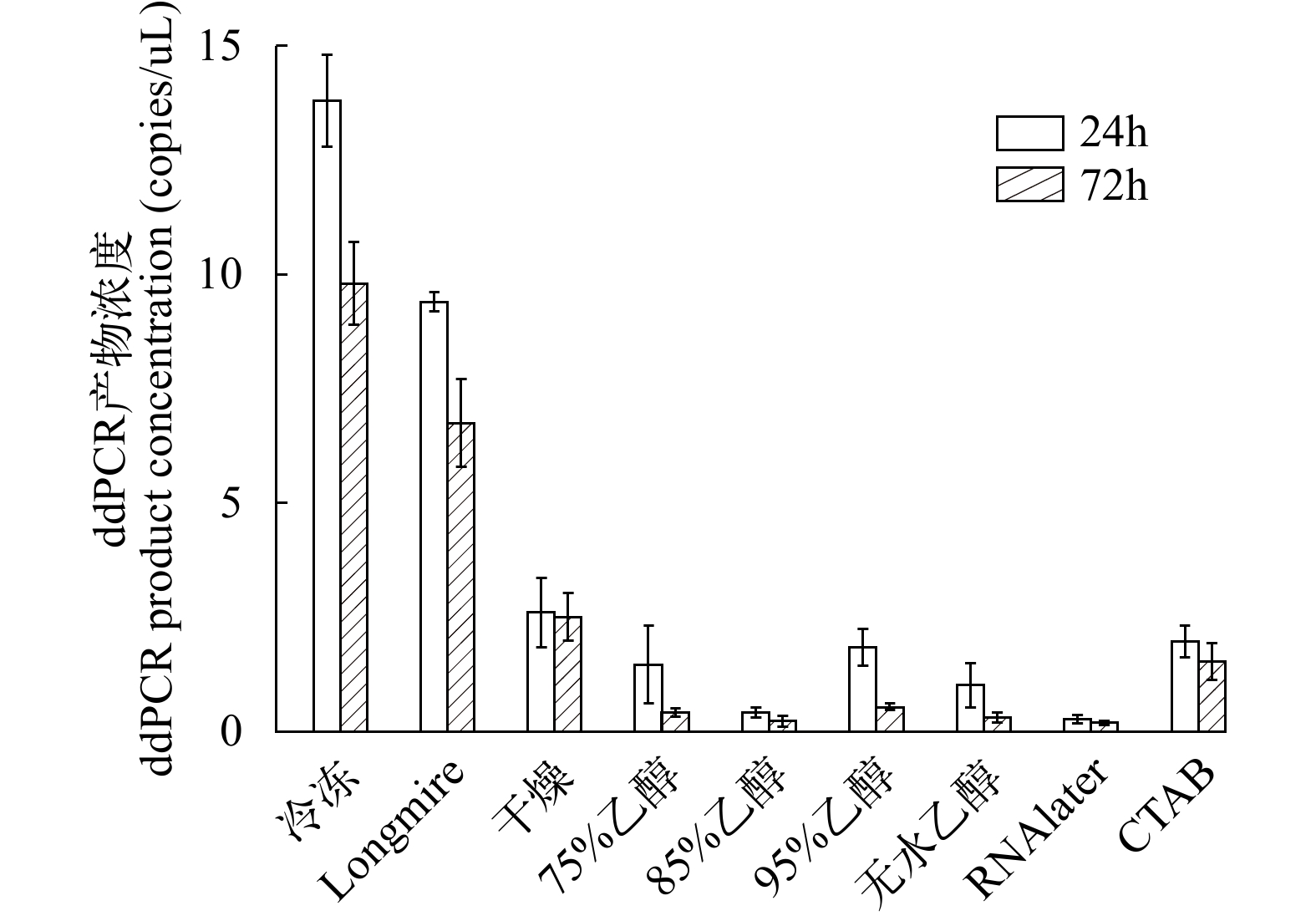
以试验水缸的水样为研究对象, 比较9种保存条件下分别保存24h和72h后获得的ddPCR产物浓度。结果发现保存24h后, –20℃冷冻保存条件下获得的ddPCR产物浓度最高, 为(13.80±1.41) copies/μL; 其次是Longmire’s buffer保存液, 为(9.40±0.21) copies/μL; 显著高于其他保存方法(P<0.05)。保存72h的样品DNA提取后, 进行ddPCR实验, 结果显示–20℃冷冻保存获得的ddPCR产物浓度最高, 利用Longmire’s buffer保存液次之。随机抽取的PCR检测结果表明ddPCR产物均为鲫的目标片段。综上所述, 滤膜保存效果最好的是–20℃冷冻保存和Longmire’s buffer保存液(图 6)。
2.5 免DNA提取PCR效果
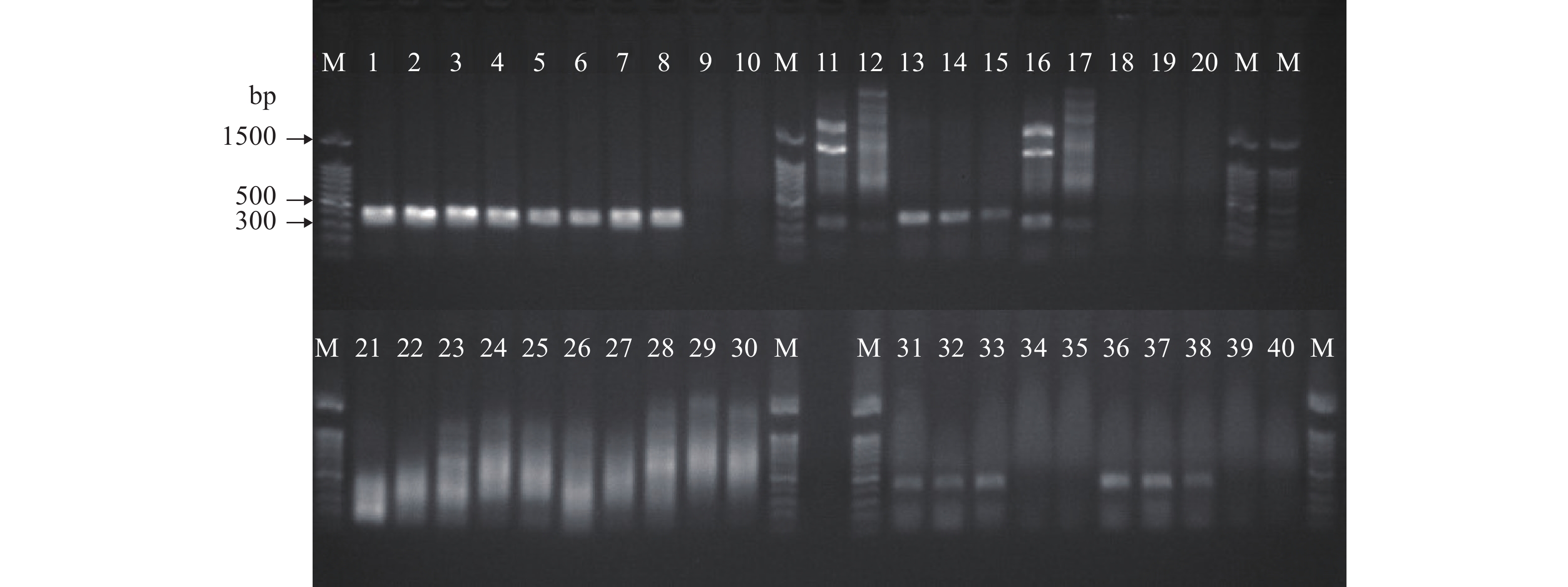
以试验水缸的水样为研究对象, 通过对PCR体系不断优化来比较4种免DNA提取PCR直扩方法的效果。结果表明, 诺唯赞高保真酶扩增效果最好, 且直接添加水样当模板的扩增效果与酒精沉淀预处理水样的效果相似。最优的反应体系为(25 μL): 2×PCR buffer为12.5 μL, dNTPs(2.5 mmol/L)为0.5 μL, AcMDB07-F和AcMDB07-R各为1 μL, DNA模板为2.5 μL, Taq酶为0.5 μL, ddH2O为7 μL。扩增产物送样测序后证明是目的片段。进一步用武汉市东湖水样进行实验, 验证了诺唯赞高保真酶的扩增条带最亮, 但诺唯赞高效酶和生工PCR直扩试剂盒也可以扩增出相应的目的片段, TaKaRa免提取试剂盒不能扩增出目标片段(图 7)。
![]() 图 7 武汉东湖水样免DNA提取PCR直扩验证1、6、11、16、21、26、31和36为实验室水样; 2、7、12、17、22、27、32和37为东湖水样; 3、8、13、18、23、28、33和38为酒精沉淀预处理实验室水样; 4、9、14、19、24、29、34和39为酒精沉淀预处理东湖水样1; 5、10、15、20、25、30、35和40为酒精沉淀预处理东湖水样2; 1—10为诺唯赞高保真酶(P505-d1); 11—20为诺唯赞高效酶(P213-AA); 21—30为TerraTM PCR Direct Polymerase Mix; 31—40为生工直接PCR扩增试剂盒; 1—5, 11—15, 21—25, 31—35水样DNA添加量为2.5 μL; 6—10, 16—20, 26—30, 36—40水样DNA添加量为9.5 μL; M为DNA Marker(100 bp Ladder)Figure 7. Validation of DNA extraction-free PCR direct amplification in water samples from Wuhan East Lake1, 6, 11, 16, 21, 26, 31, 36: Laboratory water; 2, 7, 12, 17, 22, 27, 32, 37: East Lake water; 3, 8, 13, 18, 23, 28, 33, 38: Alcohol precipitation laboratory water samples; 4, 9, 14, 19, 24, 29, 34, 39: Alcohol precipitation East Lake water sample 1; 5, 10, 15, 20, 25, 30, 35, 40: Alcohol precipitation East Lake water sample 2; 1—10: Phanta® Max Super-Fidelity DNA Polymerase; 11—20: 2xTaq Plus Master Mix II; 21—30: TerraTM PCR Direct Polymerase Mix; 31—40 Sangon® Direct PCR kit; 1—5, 11—15, 21—25 and 31—35: DNA template 2.5 μL; 6—10, 16—20, 26—30 and 36—40: DNA template 9.5 μL; M: DNA Marker(100 bp Ladder)
图 7 武汉东湖水样免DNA提取PCR直扩验证1、6、11、16、21、26、31和36为实验室水样; 2、7、12、17、22、27、32和37为东湖水样; 3、8、13、18、23、28、33和38为酒精沉淀预处理实验室水样; 4、9、14、19、24、29、34和39为酒精沉淀预处理东湖水样1; 5、10、15、20、25、30、35和40为酒精沉淀预处理东湖水样2; 1—10为诺唯赞高保真酶(P505-d1); 11—20为诺唯赞高效酶(P213-AA); 21—30为TerraTM PCR Direct Polymerase Mix; 31—40为生工直接PCR扩增试剂盒; 1—5, 11—15, 21—25, 31—35水样DNA添加量为2.5 μL; 6—10, 16—20, 26—30, 36—40水样DNA添加量为9.5 μL; M为DNA Marker(100 bp Ladder)Figure 7. Validation of DNA extraction-free PCR direct amplification in water samples from Wuhan East Lake1, 6, 11, 16, 21, 26, 31, 36: Laboratory water; 2, 7, 12, 17, 22, 27, 32, 37: East Lake water; 3, 8, 13, 18, 23, 28, 33, 38: Alcohol precipitation laboratory water samples; 4, 9, 14, 19, 24, 29, 34, 39: Alcohol precipitation East Lake water sample 1; 5, 10, 15, 20, 25, 30, 35, 40: Alcohol precipitation East Lake water sample 2; 1—10: Phanta® Max Super-Fidelity DNA Polymerase; 11—20: 2xTaq Plus Master Mix II; 21—30: TerraTM PCR Direct Polymerase Mix; 31—40 Sangon® Direct PCR kit; 1—5, 11—15, 21—25 and 31—35: DNA template 2.5 μL; 6—10, 16—20, 26—30 and 36—40: DNA template 9.5 μL; M: DNA Marker(100 bp Ladder)3. 讨论
研究表明较高的eDNA浓度可以提高物种丰富度检测, 并提高不同eDNA研究的一致性[22, 40]。本研究的实验结果表明, eDNA实验中捕获方法和DNA提取方法的选择对最终ddPCR产物浓度有显著影响。基于不同捕获方法和DNA提取方法的组合实验确定了最佳的eDNA捕获和提取的方法, 即通过0.22 μm孔径的混合纤维素膜捕获水样中的eDNA, 然后使用Qiagen DNeasy PowerWater kit进行DNA的提取。与最差的eDNA组合方案(聚四氟乙烯膜抽滤和酚氯仿异戊醇法提取DNA)相比, ddPCR产物浓度增加了近22倍。因此, 本研究表明了eDNA样本处理方案优化的重要性, 对eDNA标准操作流程进行优化和标准化确保了各研究之间的可比性, 同时促进eDNA技术在水生生物物种多样性监测和保护中的应用。
3.1 滤膜筛选和eDNA提取方法综合评价
本研究对6种滤膜进行了比较, 结果表明混合纤维素膜在捕获eDNA总产量上显著优于同一孔径的其他5种滤膜(图 3)。该结果与Liang等[24]和Majaneva等[41]的研究结果一致, Liang等[24]研究了不同材质滤膜对DNA捕获效率的影响, 发现DNA对不同材质的滤膜有着不同的结合能力, 混合纤维素膜捕获的eDNA总产量最高, 其次是聚偏氟乙烯膜(本研究未测)和聚碳酸酯膜, 进一步验证了混合纤维素膜的捕获效率。尽管本研究结果与Liang等[24]一致, 但是他们的研究对象是纯化的DNA, 本研究则是水样中的eDNA, 既包括胞外和胞内DNA, 还包括一些小的组织碎片或粪便, 包括了水样中所有形式的DNA, 更适用于野外eDNA采样。
DNA与各种滤膜类型的不同结合能力可能与滤膜本身的特性有关。例如, 滤膜分为深层过滤(将细胞碎片和DNA等成分保留在滤膜表面和滤膜基质内, 例如: 玻璃纤维素膜、硝酸纤维素膜和混合纤维素膜)和表面过滤(将细胞碎片和DNA等成分截留在滤膜表面, 例如: 聚碳酸酯膜)[18, 42]。这可以用来解释为什么混合纤维素膜(二醋酸纤维素和硝酸纤维素的混合物)和硝酸纤维素膜捕获DNA效果比其他滤膜好, 一方面可能是DNA被截留在滤膜基质内, 并不仅仅是滤膜表面。另一方面可能是硝酸纤维素膜本身固有的高DNA和蛋白质结合能力[43, 44]。
本研究对5种不同的DNA提取方法进行了比较研究, 发现Qiagen DNeasy PowerWater kit提取获得的ddPCR产物浓度最高, Qiagen DNeasy Blood & Tissue kit和酚氯仿异戊醇法提取次之。在未对Qiagen DNeasy PowerWater kit进行比较的情况下, Hinlo等[18]和Deiner等[19]研究认为 Qiagen DNeasy Blood & Tissue kit的DNA提取效果最佳, 本研究结果也进一步证实了他们的结论。Qiagen DNeasy Blood & Tissue kit提取DNA时, 滤膜在裂解缓冲液和蛋白酶K中孵育几小时, 主要依靠生化方法来裂解细胞。而本研究中的Qiagen DNeasy PowerWater kit提取DNA主要是依靠机械作用-打珠[19], 同时配合含有抑制因子去除技术的强力裂解试剂, 打碎滤膜同时辅助裂解细胞, 可能会使滤膜基质内释放出更多的核酸。另外也有研究表明, 与DNA提取试剂盒相比, 酚氯仿异戊醇法可以获得更多的eDNA产物[19, 23]。虽然酚氯仿异戊醇法价格低廉, 但是需要提前准备和处理有毒化学品, 并且该提取方法耗时较长、有毒、需要通风橱等弊端。相比之下, DNA试剂盒使用简单方便, 且本研究发现DNA试剂盒提取的ddPCR产物更多。对于极低密度物种的检测应用, 较低的eDNA回收产量可能意味着检测失败, 在这种情况下, Qiagen DNeasy PowerWater kit的高eDNA回收产量能在eDNA应用中发挥重要作用。
通过对eDNA捕获和提取方法结果进行双因素方差分析, 表明不同的eDNA捕获和提取方法之间存在显著的交互作用(F=20.59, P<0.001)。这一研究结果与Eichmiller 等[20]和Renshaw等[23]研究一致, 但是这些研究使用的是材料种类相对较少、孔径不一的滤膜, 由于滤膜孔径大小不一, 所以同样体积的水过滤后的DNA产量会存在差异, 因此直接使用最终的ddPCR产物浓度来评估滤膜的性能就不具有可比性。而本研究是对同一孔径的不同材质滤膜进行了比较, 保证了除材质外其他条件的一致性, 可以直接通过对eDNA产量进行比较来评估各种材料滤膜的性能。不同滤膜和不同DNA提取方法的交互效应分析显示混合纤维素膜总体捕获效果最好, 其次是硝酸纤维素膜, 这与Majaneva等[41]研究结果一致。除DNA产量之外, 滤膜的流速也是eDNA实验考虑的另一重要因素, 具有较高流速的滤膜可以显著缩短过滤时间和相关的人工成本。对于较为浑浊的水体, 抽滤时间可能是一个关键因素, 长时间的抽滤可能会导致eDNA降解。本研究对相同体积水样的六种滤膜进行比较研究, 发现玻璃纤维素膜抽滤时间最短, 其次为聚碳酸酯膜、混合纤维膜、硝酸纤维膜, 尼龙膜和聚四氟乙烯膜的抽滤时间最长。综上所述, 本研究发现, 使用Qiagen DNeasy PowerWater Kit提取的混合纤维素膜的组合为最优组合。但是, eDNA样本处理方法的选择往往会因研究目标、个人偏好性、易用性等方面而有所不同, 本研究结果可以为后续eDNA技术和方法研究提供参考。如果对大量浑浊水样进行抽滤, 则玻璃纤维素膜过滤或更大孔径的硝酸纤维素膜可能更适合。对于小体积水样抽滤, 混合纤维素膜和Qiagen DNeasy Blood & Tissue kit组合可能在价格、时间成本和人工成本方面优于其他组合, 并且捕获eDNA产量较高。
3.2 滤膜保存方法评价
本研究通过对常用的滤膜保存方法(干燥、冷冻、RNAlater、CTAB buffer、Longmire’s buffer及不同浓度酒精保存)获得的ddPCR产物浓度进行研究, 结果发现–20℃冷冻保存和Longmire’s buffer保存效果显著优于其余的保存方法, 这一研究结果与Mauvisseau等[45]、Renshaw等[23]和Kumar等[46]的研究结果一致。Longmire’s buffer保存液的保存效果, 大大降低了常温条件下DNA降解速率, 使eDNA在常温下也可以短时间(不超过24h)储存, 降低采样成本, 成功实现了环境温度保存。这一发现使eDNA在一些偏远地区或热带地区进行研究成为了可能。有研究表明, 在乙醇中储存6d的滤膜eDNA浓度并没有显著差异[36], 本研究结果与其一致。但是无水乙醇保存24h和72h后滤膜的ddPCR产物浓度都显著低于–20℃冷冻保存和Longmire’s buffer保存(P<0.001; 图 6)。综上所述, 建议在无冷冻条件情况下用Longmire’s buffer对滤膜进行保存, 这一建议与早期研究发现是一致的。
3.3 免DNA提取PCR直扩
本研究对免DNA提取PCR直扩实验进行了初步探究, 对TaKaRa免抽提试剂盒(TerraTM PCR Direct Polymerase Mix)、诺唯赞高保真酶(P505-d1)、诺唯赞高效酶(P213-AA)和生工直接PCR扩增试剂盒4种免DNA提取PCR直扩方法的效果进行比较。研究结果发现, PCR扩增效果并不会随着模板量的增大而变好, 一方面可能是模板量增多, 蛋白含量较高会影响PCR反应体系; 另一原因可能是水样中含有抑制剂, 随着模板量增大抑制剂浓度增高, 从而导致扩增效果不好。实验结果表明, 诺唯赞高保真酶的扩增效果最好, 显著优于其余3种直扩方法。最优反应体系为(25 μL): 2×PCR buffer为12.5 μL, dNTPs(2.5 mmol/L)为0.5 μL, AcMDB07-F和AcMDB07-R各为1 μL, DNA模板为2.5 μL, Taq酶为0.5 μL, ddH2O为7 μL。因此, 建议用诺唯赞高保真酶进行PCR直扩实验来实现免DNA提取。
4. 结论
eDNA方法的选择除了成本和方便使用之外, 还应该考虑效率、可靠性和可比性。本研究进一步证明了eDNA捕获、提取和保存的选择可以显著影响DNA产量。通过使用同一孔径的不同材质滤膜, 能够直接对相同DNA提取的不同滤膜的eDNA产量进行比较。因为研究环境的物理和化学特性并不相同[47—49], 通用的eDNA捕获和提取方法可能不适用所有的eDNA研究。但是基于本研究的结果, 对于淡水生态系统较清澈水体, 建议使用混合纤维素膜对水样eDNA进行抽滤来捕获eDNA, 然后组合Qiagen DNeasy PowerWater kit进行DNA提取。对于在野外无冷藏条件情况下, 建议使用Longmire’s buffer保存液将滤膜短时间(不超过24h)常温保存, 寄回实验室进行DNA提取。对于免DNA提取初步研究结果建议采用诺唯赞高保真酶进行PCR直扩。
-
图 5 滤膜和eDNA提取方法组合比较
a. 同一滤膜类型在不同DNA提取方法下获得的ddPCR产物浓度; b. DNA提取方法的主效应图, 即不考虑滤膜, 所有样品用不同DNA提取方法获得的ddPCR产物浓度; c. 滤膜类型的主效应图, 即不考虑提取方法, 所有样品用不同类型滤膜过滤获得的ddPCR产物浓度; d. 同一提取方法在不同滤膜过滤情况下获得的ddPCR产物浓度
Figure 5. The comparison of cross-combination of filters and DNA extraction methods
a. The ddPCR concentration obtained by the same filter under different DNA extraction methods; b. The main effect of the DNA extraction methods. The ddPCR concentration obtained by different DNA extraction methods for all samples without considering the filters; c. The main effect of the filter types, The ddPCR concentration obtained by different filter types for all samples without considering the DNA extraction methods; d. The ddPCR concentration obtained by the same extraction method under different filters
图 7 武汉东湖水样免DNA提取PCR直扩验证
1、6、11、16、21、26、31和36为实验室水样; 2、7、12、17、22、27、32和37为东湖水样; 3、8、13、18、23、28、33和38为酒精沉淀预处理实验室水样; 4、9、14、19、24、29、34和39为酒精沉淀预处理东湖水样1; 5、10、15、20、25、30、35和40为酒精沉淀预处理东湖水样2; 1—10为诺唯赞高保真酶(P505-d1); 11—20为诺唯赞高效酶(P213-AA); 21—30为TerraTM PCR Direct Polymerase Mix; 31—40为生工直接PCR扩增试剂盒; 1—5, 11—15, 21—25, 31—35水样DNA添加量为2.5 μL; 6—10, 16—20, 26—30, 36—40水样DNA添加量为9.5 μL; M为DNA Marker(100 bp Ladder)
Figure 7. Validation of DNA extraction-free PCR direct amplification in water samples from Wuhan East Lake
1, 6, 11, 16, 21, 26, 31, 36: Laboratory water; 2, 7, 12, 17, 22, 27, 32, 37: East Lake water; 3, 8, 13, 18, 23, 28, 33, 38: Alcohol precipitation laboratory water samples; 4, 9, 14, 19, 24, 29, 34, 39: Alcohol precipitation East Lake water sample 1; 5, 10, 15, 20, 25, 30, 35, 40: Alcohol precipitation East Lake water sample 2; 1—10: Phanta® Max Super-Fidelity DNA Polymerase; 11—20: 2xTaq Plus Master Mix II; 21—30: TerraTM PCR Direct Polymerase Mix; 31—40 Sangon® Direct PCR kit; 1—5, 11—15, 21—25 and 31—35: DNA template 2.5 μL; 6—10, 16—20, 26—30 and 36—40: DNA template 9.5 μL; M: DNA Marker(100 bp Ladder)
-
[1] Butchart S H M, Walpole M, Collen B, et al. Global biodiversity: indicators of recent declines [J]. Science, 2010, 328(5982): 1164-1168. doi: 10.1126/science.1187512
[2] Wang P, Yan Z, Yang S, et al. Environmental DNA: an emerging tool in ecological assessment [J]. Bulletin of Environmental Contamination and Toxicology, 2019, 103(5): 651-656. doi: 10.1007/s00128-019-02720-z
[3] Bohmann K, Evans A, Gilbert M, et al. Environmental DNA for wildlife biology and biodiversity monitoring [J]. Trends in Ecology & Evolution, 2014, 29(29): 358-367.
[4] 马鸿娟, Stewart Kathryn, 马利民, 等. 环境DNA及其在水生生态系统保护中的应用 [J]. 生态学杂志, 2016, 35(2): 244-251. Ma H J, Stewart K, Ma L M, et al. Environmental DNA and its application in protecting aquatic ecosystems [J]. Chinese Journal of Ecology, 2016, 35(2): 244-251.
[5] Jeunen G J, Knapp M, Spencer H G, et al. Species-level biodiversity assessment using marine environmental DNA metabarcoding requires protocol optimization and standardization [J]. Ecology and Evolution, 2019, 9(3): 1323-1335. doi: 10.1002/ece3.4843
[6] Barnes M A, Turner C R. The ecology of environmental DNA and implications for conservation genetics [J]. Conservation Genetics, 2016, 17(1): 1-17. doi: 10.1007/s10592-015-0775-4
[7] Yang J, Zhang X. eDNA metabarcoding in zooplankton improves the ecological status assessment of aquatic ecosystems [J]. Environment International, 2020(134): 105230.
[8] Cilleros K, Valentini A, Allard L, et al. Unlocking biodiversity and conservation studies in high-diversity environments using environmental DNA (eDNA): A test with Guianese freshwater fishes [J]. Molecular Ecology Resources, 2019, 19(1): 27-46. doi: 10.1111/1755-0998.12900
[9] Rees H C, Maddison B C, Middleditch D J, et al. Review: The detection of aquatic animal species using environmental DNA - a review of eDNA as a survey tool in ecology [J]. Journal of Applied Ecology, 2014, 51(5): 1450-1459. doi: 10.1111/1365-2664.12306
[10] Mahon A R, Jerde C L, Galaska M, et al. Validation of eDNA surveillance sensitivity for detection of asian carps in controlled and field experiments [J]. PLoS One, 2013, 8(3): e58316. doi: 10.1371/journal.pone.0058316
[11] Xu N, Zhu B, Shi F, et al. Monitoring seasonal distribution of an endangered anadromous sturgeon in a large river using environmental DNA [J]. Die Naturwissenschaften, 2018, 105(11-12): 62. doi: 10.1007/s00114-018-1587-4
[12] Takahara T, Minamoto T, Yamanaka H, et al. Estimation of fish biomass using environmental DNA [J]. PLoS One, 2012, 7(4): e35868. doi: 10.1371/journal.pone.0035868
[13] Kelly R P, Port J A, Yamahara K M, et al. Using environmental dna to census marine fishes in a large mesocosm [J]. PLoS One, 2014, 9(1): e86175. doi: 10.1371/journal.pone.0086175
[14] Jerde C L, Wilson E A, Dressler T L. Measuring global fish species richness with eDNA metabarcoding [J]. Molecular Ecology Resources, 2019, 19(1): 19-22. doi: 10.1111/1755-0998.12929
[15] 舒璐, 林佳艳, 徐源, 等. 基于环境DNA宏条形码的洱海鱼类多样性研究 [J]. 水生生物学报, 2020, 44(5): 7. Shu L, Lin J Y, Xu Y, et al. Investigating the fish diversity in Erhai Lake based on environmental dna metabarcoding [J]. Acta Hydrobiologica Sinica, 2020, 44(5): 7.
[16] Ahn H, Kume M, Terashima Y, et al. Evaluation of fish biodiversity in estuaries using environmental DNA metabarcoding [J]. PLoS One, 2020, 15(10): e0231127. doi: 10.1371/journal.pone.0231127
[17] Thomsen P F, Willerslev E. Environmental DNA-An emerging tool in conservation for monitoring past and present biodiversity [J]. Biological Conservation, 2015(183): 4-18.
[18] Hinlo R, Gleeson D, Lintermans M, et al. Methods to maximise recovery of environmental DNA from water samples [J]. PLoS One, 2017, 12(6): e0179251. doi: 10.1371/journal.pone.0179251
[19] Deiner K, Walser J C, Mächler E, et al. Choice of capture and extraction methods affect detection of freshwater biodiversity from environmental DNA [J]. Biological Conservation, 2015(183): 53-63.
[20] Eichmiller J J, Miller L M, Sorensen P W. Optimizing techniques to capture and extract environmental DNA for detection and quantification of fish [J]. Molecular Ecology Resources, 2016, 16(1): 56-68. doi: 10.1111/1755-0998.12421
[21] Spens J, Evans A R, Halfmaerten D, et al. Comparison of capture and storage methods for aqueous macrobial eDNA using an optimized extraction protocol: advantage of enclosed filter [J]. Methods in Ecology and Evolution, 2016, 8(5): 635-645.
[22] Djurhuus A, Port J, Closek C J, et al. Evaluation of Filtration and DNA extraction methods for environmental DNA biodiversity assessments across multiple trophic levels [J]. Frontiers in Marine Science, 2017(4): 314.
[23] Renshaw M A, Olds B P, Jerde C L, et al. The room temperature preservation of filtered environmental DNA samples and assimilation into a phenol-chloroformisoamyl alcohol DNA extraction [J]. Molecular Ecology Resources, 2015, 15(1): 168-176. doi: 10.1111/1755-0998.12281
[24] Liang Z, Keeley A. Filtration recovery of extracellular DNA from environmental water samples. Environmental Science & Technology, 2013. 47(16): 9324-9331.
[25] 赵新, 兰青阔, 陈锐, 等. 应用微滴数字PCR技术快速检测食用菌中沙门氏菌 [J]. 食品与生物技术学报, 2017, 36(3): 315-321. doi: 10.3969/j.issn.1673-1689.2017.03.014 Zhao X, Lan Q K, Chen R, et al. Rapid detection of salmonella spp. in edible fungi by droplet digital PCR [J]. Journal of Food Science and Biotechnology, 2017, 36(3): 315-321. doi: 10.3969/j.issn.1673-1689.2017.03.014
[26] Doi H, Takahara T, Minamoto T, et al. Droplet digital polymerase chain reaction (PCR) outperforms real-time PCR in the detection of environmental DNA from an invasive fish species [J]. Environmental Science & Technology, 2015, 49(9): 5601-5608.
[27] Mauvisseau Q, Davy-Bowker J, Bulling M, et al. Improving detection capabilities of a critically endangered freshwater invertebrate with environmental DNA using digital droplet PCR [J]. BioRxiv, 2019,DOI: 10.1101/661447.
[28] Wood S A, Pochon X, Laroche O, et al. A comparison of droplet digital polymerase chain reaction (PCR), quantitative PCR and metabarcoding for species-specific detection in environmental DNA [J]. Molecular Ecology Resources, 2019, 19(6): 1407-1419. doi: 10.1111/1755-0998.13055
[29] Whale AS, Huggett J F, Simon C, et al. Comparison of microfluidic digital PCR and conventional quantitative PCR for measuring copy number variation [J]. Nucleic Acids Research, 2012, 40(11): e82. doi: 10.1093/nar/gks203
[30] Hindson B J, Ness K D, Masquelier D A, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number [J]. Analytical Chemistry, 2011, 83(22): 8604-8610. doi: 10.1021/ac202028g
[31] Deiner K, Altermatt F. Transport distance of invertebrate environmental DNA in a natural river [J]. PLoS One., 2014, 9(2): e88786. doi: 10.1371/journal.pone.0088786
[32] Costas B A, Mcmanus G, Doherty M, et al. Use of species-specific primers and PCR to measure the distributions of planktonic ciliates in coastal waters [J]. Limnology and Oceanography Methods, 2007, 5(6): 163-173. doi: 10.4319/lom.2007.5.163
[33] Aljanabi S M, Martinez I. Universal and rapid salt-extraction of high quality gnomic DNA for PCR-based techniques [J]. Nucleic Acids Research, 1997, 25(22): 4692-4693. doi: 10.1093/nar/25.22.4692
[34] Coyne K J, Handy S M, Demir E, et al. Improved quantitative real-time PCR assays for enumeration of harmful algal species in field samples using an exogenous DNA reference standard [J]. Limnology and Oceanography Methods, 2005, 3(9): 381-391. doi: 10.4319/lom.2005.3.381
[35] Longmire J L, Maltbie M, Baker R J. Use of 'Lysis Buffer' in DNA isolation and its implication for museum collections [J]. Occasional Papers Museum of Texas Tech University, 1997(163): 1-3.
[36] Minamoto T, Naka T, Moji K, et al. Techniques for the practical collection of environmental DNA: filter selection, preservation, and extraction [J]. Limnology, 2016, 17(1): 23-32. doi: 10.1007/s10201-015-0457-4
[37] Jerde C L, Chadderton W L, Mahon A R, et al. Detection of Asian carp DNA as part of a Great Lakes basin-wide surveillance program [J]. Canadian Journal of Fisheries and Aquatic Sciences, 2013, 70(4): 522-526. doi: 10.1139/cjfas-2012-0478
[38] Jerde C L, Mahon A R, Chadderton W L, et al. “Sight-unseen” detection of rare aquatic species using environmental DNA [J]. Conservation Letters, 2011, 4(2): 150-157. doi: 10.1111/j.1755-263X.2010.00158.x
[39] Bylemans J, Gleeson D M, Hardy C M, et al. Toward an ecoregion scale evaluation of eDNA metabarcoding primers: A case study for the freshwater fish biodiversity of the Murray-Darling Basin (Australia) [J]. Ecology and Evolution, 2018, 8(17): 8697-8712. doi: 10.1002/ece3.4387
[40] Goldberg C S, Strickler K M, Pilliod D S. Moving environmental DNA methods from concept to practice for monitoring aquatic macroorganisms [J]. Biological Conservation, 2015(183): 1-3. doi: 10.1016/j.biocon.2014.11.040
[41] Majaneva M, Diserud O H, Eagle S H C, et al. Environmental DNA filtration techniques affect recovered biodiversity [J]. Scientific Reports, 2018, 8(1): 4682. doi: 10.1038/s41598-018-23052-8
[42] 汪功伟, 李浔. 深层过滤技术在生物制药工艺中的运用 [J]. 中国化工贸易, 2015(15): 151-151. Wang G W, Li X. Application of deep filtration technology in biopharmaceutical process [J]. China Chemical Trade, 2015(15): 151-151.
[43] Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications [J]. Proceedings of the National Academy of Sciences, 1979, 76(9): 4350-4354. doi: 10.1073/pnas.76.9.4350
[44] Thornton D J, Carlstedt I, Sheehan J K. Identification of glycoproteins on nitrocellulose membranes and gels [J]. Molecular Biotechnology, 1996, 5(2): 171. doi: 10.1007/BF02789065
[45] Mauvisseau Q, Halfmaerten D, Sabrina N, et al. Effects of preservation strategies on environmental DNA detection and quantification using ddPCR [J]. Environmental DNA, 2021, 3(4): 815-822. doi: 10.1002/edn3.188
[46] Kumar G, Eble J E, Gaither M R. A practical guide to sample preservation and pre-PCR processing of aquatic environmental DNA [J]. Molecular Ecology Resources, 2020, 20(1): 29-39. doi: 10.1111/1755-0998.13107
[47] Goldberg C S, Turner C R, Deiner K, et al. Critical considerations for the application of environmental DNA methods to detect aquatic species [J]. Methods in Ecology and Evolution, 2016, 7(11): 1299-1307. doi: 10.1111/2041-210X.12595
[48] Deiner K, Bik H M, Machler E, et al. Environmental DNA metabarcoding: Transforming how we survey animal and plant communities [J]. Molecular Ecology, 2017, 26(21): 5872-5895. doi: 10.1111/mec.14350
[49] Buxton A S, Groombridge J J, Griffiths R A. Is the detection of aquatic environmental DNA influenced by substrate type [J]? PLoS One, 2017, 12(8): e0183371. doi: 10.1371/journal.pone.0183371
-
期刊类型引用(4)
1. 杨方典,张鹏,罗帅,熊雄,熊杰,缪炜. 基于宏基因组的西藏拉鲁湿地原生生物多样性研究. 水生态学杂志. 2025(01): 81-89 .  百度学术
百度学术
2. 刘洋,应方,杨俊,王焕英,洪文杰,王庭璋. 鱼类环境DNA保存条件的比较研究. 生物学杂志. 2024(03): 114-120 .  百度学术
百度学术
3. 许沛璠,杨美琳,庞婉婷,马静,汪福顺,陈学萍. 新安江水库浮游植物的鉴定及群落结构变化. 生态学杂志. 2024(10): 3160-3169 .  百度学术
百度学术
4. 李晨虹,凌岚馨,谭娟,林晓龙,王辉,孙冰皎,李曌. 环境DNA技术在水生生物监测中的挑战、突破和发展前景. 上海海洋大学学报. 2023(03): 564-574 .  百度学术
百度学术
其他类型引用(3)



 下载:
下载: