GENETIC DIVERSITY OF THE PRENANT’S SCHIZOTHORACIN (SCHIZOTHORAX PRENANTI) BASED ON PARTIAL MTDNA CYTB SEQUENCES
-
摘要: 研究测定了长江上游4个齐口裂腹鱼(Schizothorax prenanti)野生群体(重庆巫溪、重庆城口、四川雅安、四川阿坝)共104个个体的线粒体Cyt b基因部分序列, 以探讨齐口裂腹鱼野生群体的遗传多样性和遗传结构。结果表明: 在104个个体Cyt b序列中共检测到43个多态性位点, 25个单倍型。4个齐口裂腹鱼群体的单倍型多样性介于0.704—0.884, 核苷酸多样性介于0.007—0.012。群体间Kimura双参数遗传距离介于0.008—0.017, 其中四川雅安群体与四川阿坝群体间遗传距离最近, 基因交流频繁。重庆城口群体与四川雅安群体间遗传距离最远, 基因交流受阻。AMOVA分析表明, 齐口裂腹鱼的遗传分化主要来自群体内部, 且组群间、组群内群体间和群体内存在显著的遗传分化。中性检验得到Tajima’s D和 Fu’s Fs的值不显著, 且歧点分布图呈多峰, 表明长江上游4个齐口裂腹鱼野生群体未经历过种群扩张。研究旨为齐口裂腹鱼野生资源保护提供必要参考意见, 同时为齐口裂腹鱼种质资源合理开发和利用提供理论依据。Abstract: Prenant’s schizothoracin, Schizothorax prenanti (Tchang), a valuable and economic fish, distributes in the main streams and tributaries of the upper reaches of the Yangtze River in China. To assess genetic diversity and gene-tic structure of the wild populations of S. prenanti, we amplified partial Mitochondrial Cyt b gene sequences from 104 individuals of the four wild populations of S. pregnant (Wuxi, Chengkou, Ya’an and A’ba) from the upper reaches of the Yangtze River. The results showed that 43 polymorphic nucleotide sites and 25 haplotypes were detected among the Cyt b sequences of 104 individuals. The haplotype diversity was ranged from 0.704 to 0.884, and nucleotide diversity index was ranged from 0.007 to 0.012 among the four populations. Kimura 2-parameter distance was ranged from 0.008 and 0.017. The genetic distance between Ya’an and A’ba population was the smallest and the gene exchange between the two populations was frequent. The largest genetic distance was observed between Ya’an and Chengkou population, which might be explained by the little gene exchange between the two populations. AMOVA analysis showed that the genetic differentiation of S. prenanti was mainly occurred within populations variation. There were significant genetic differentiations between the groups, among populations within groups, and within populations. The results of neutrality test showed that the values of Tajima’s D and Fu’s Fs were not significant and the mismatch distribution was multimodal, indicating that the four populations of S. prenanti from the upper reaches of the Yangtze River had not experienced population expansion. This study provides valuable knowledge for protecting Schizothorax prenanti and theore-tic information for a rational utilization of the germplasm resources.
-
Keywords:
- Schizothorax prenanti /
- Cyt b /
- Genetic structure /
- Genetic diversity
-
齐口裂腹鱼(Schizothorax prenanti)俗称雅鱼、洋鱼、齐口细鳞鱼, 属鲤形目、鲤科、裂腹鱼亚科、裂腹鱼属, 主要分布于我国长江上游的金沙、岷江、大渡河、青衣江及乌江下游等水域, 在长江上游淡水生态系统的食物链中具有重要的地位[1,2]。近年来, 由于自然环境恶化及人为活动的不利影响, 如过度捕捞、修建电站、水体污染等导致齐口裂腹鱼野生资源遭到严重破坏, 其天然资源量逐渐减少, 种群数量急剧下降[3]。因此, 分析齐口裂腹鱼不同群体的遗传多样性和遗传结构, 对其资源的保护有着重要的理论价值和实际意义。
迄今对齐口裂腹鱼的研究多集中于生物学特性、营养与生长、繁殖和疾病防治等方面[4—7], 而对于其遗传资源的调查、评估, 及种群结构的研究较少, 研究者仅采用AFLP和mtDNA控制区标记分析了位于四川境内的青衣江、大渡河、雅砻江上游的木里河及伍须湖齐口裂腹鱼群体的遗传多样性[8—10], 而长江上游重庆段干流及支流的野生群体群体结构情况等未见报道。鱼类mtDNA具有分子较小、为共价闭合环状双链的DNA分子、严格的母系遗传、快速的进化速度等特点[11,12], 因此被广泛用于鱼类亲缘关系、种群遗传分化、遗传多样性和种群历史动态等研究[13—16]。在线粒体的众多基因中, 细胞色素b (Cytochrome b, Cyt b)基因进化速度适中, 转换或颠换比例较大, 被广泛应用于系统发育、遗传多样性、物种鉴定研究中[17—20]。本研究以重庆和四川齐口裂腹鱼野生群体为研究对象, 基于Cyt b分析齐口裂腹鱼种群遗传多样性和种群遗传结构, 以期为齐口裂腹鱼种群资源的保护和合理开发利用提供理论依据。
1. 材料与方法
1.1 实验材料与DNA提取
在2014年3月至2015年5月期间, 收集了重庆巫溪和城口、四川雅安和阿坝州4个齐口裂腹鱼野生群体(表 1), 剪取鳍条用无水乙醇固定, 置于–20℃冰箱低温保存。基因组DNA采用动物基因组DNA抽提试剂盒(生工生物工程股份有限公司, 上海)提取, 使用核酸蛋白定量仪(Eppendorf, BioPhotome-ter Plus, 德国)检测所提DNA浓度与质量, 并用1%琼脂糖电泳检测其完整性。DNA样品稀释成30 ng/mL的终浓度, 保存于–20℃备用。
表 1 齐口裂腹鱼样本采集信息Table 1. Sampling information of S. prenanti群体 Population 河流 River 经纬度 Longitude and Latitude 采样时间 Time 样本数 Sample size 重庆巫溪WX 大宁河 +109.31, +31.35 2014.5— 2015.5 16 重庆城口CK 任河 +108.15, +31.37 2014.7—9 35 四川雅安YA 大渡河 +101.55, +28.51 2015.4 24 四川阿坝州AB 脚木足河 +102.22, +31.89 2014.3—4 29 1.2 PCR扩增与测序
根据齐口裂腹鱼mtDNA全序列[21](GenBank序列号: NC_023829)采用Primer 5.0软件设计Cyt b基因的引物, 并由英俊生物技术服务有限公司合成。上游引物和下游引物分别为: CYF: 5′-CGTGAACT ACGGCTGACT-3′; CYR: 5′-TAGGAACTGGGTG ATTGG-3′。
PCR扩增体系为50 μL, 包括10×PCR buffer (with Mg2+) 5 μL、dNTPs (10 mmol/L) 4 μL、Taq酶(5 U/μL) 0.5 μL、上游及下游引物(10 μmol/L)各2.5 μL、模板DNA(30 ng/μL)2.5 μL, 补充无菌水至50 μL。所需试剂购自大连TaKaRa生物工程有限公司。PCR扩增程序为: 94℃预变性5min; 94℃变性30s, 55℃退火30s, 72℃延伸45s, 反应25个循环; 72℃延伸10min, 15℃保存。扩增产物用1%琼脂糖凝胶电泳检测, 送上海英骏生物技术有限公司进行PCR扩增产物的纯化、回收与单向测序, 测序引物为扩增引物。
1.3 序列整理与数据分析
采用BioEdit 7.0软件[22]和Clustal X 1.81软件[23]对测序结果进行编辑和同源比对。群体序列遗传多样性参数, 包括多态性位点数(S)、单倍型数(H)、单倍型多样性(Hd)、核苷酸多样性(π)、平均核苷酸差异数(K)和Tajima’s D值等由DnaSP 5.0[24]软件获得。
利用软件Arlequin 3.5[25]进行核苷酸组成统计、分子方差分析(AMOVA)和遗传分化指数(Φst)计算。使用Nm= (1–Φst)/(2Φst)计算两两群体间的基因流。利用Network 4.6.1.0软件[26]构建单倍型的简约中介(Reduced-Median, MJ)网络图, 用以检测单倍型之间的进化关系。利用MEGA 5.1软件[27]计算群体间的Kimura双参数模型(Kimura 2 Parameter, K2P)遗传距离, 并构建邻接(Neighbor-Joinning, NJ)进化树和最大简约(Maximum Parsimony, MP)进化树。使用Arlequin 3.5进行Tajima’s D检验[28]和Fu’s Fs检验[29], 同时结合核苷酸歧点分布分析(Mismatch distributions)来检验群体历史动态, 以确定是否存在瓶颈效应或群体扩张[30]。对显著性检验的P值进行Bonferonni校正[31]。
2. 结果
2.1 序列变异特征
共测序获得104尾齐口裂腹鱼的Cyt b基因部分序列, 序列长750 bp。104条序列中共检测到68个变异位点, 变异比例为9.86%。其中单一信息位点有33个, 简约信息位点35个。变异均为转换或颠换, 转换与颠换的比值为54.33。4个种群所有个体序列中的A、T、C、G碱基组成比例为25.50%、29.70%、27.80%以及17.00%, A+T(55.20%)含量明显高于G+C(44.80%)含量, 其中G含量最低(17.00%), 表现出明显的反G偏倚特点。
2.2 单倍型分布及遗传关系
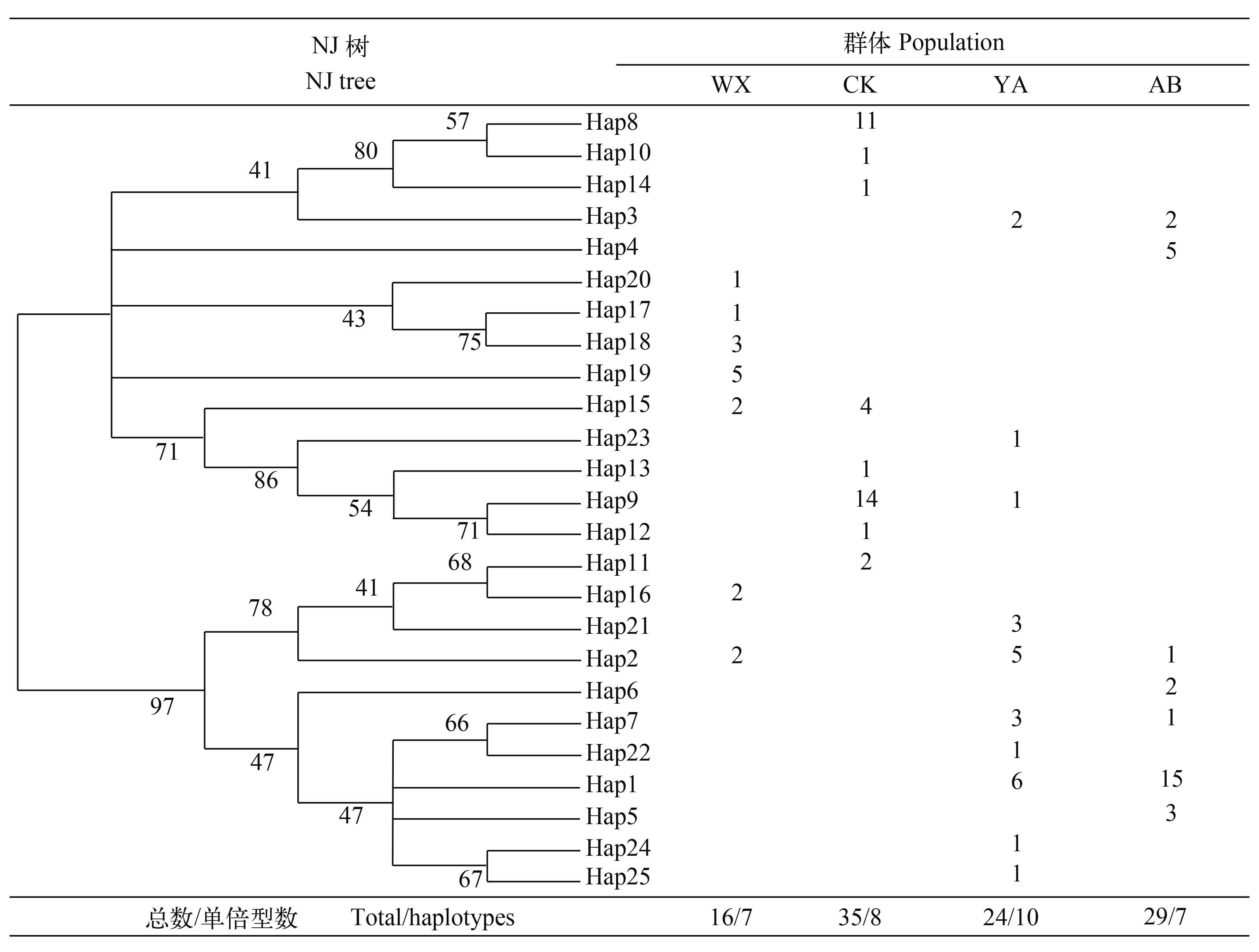
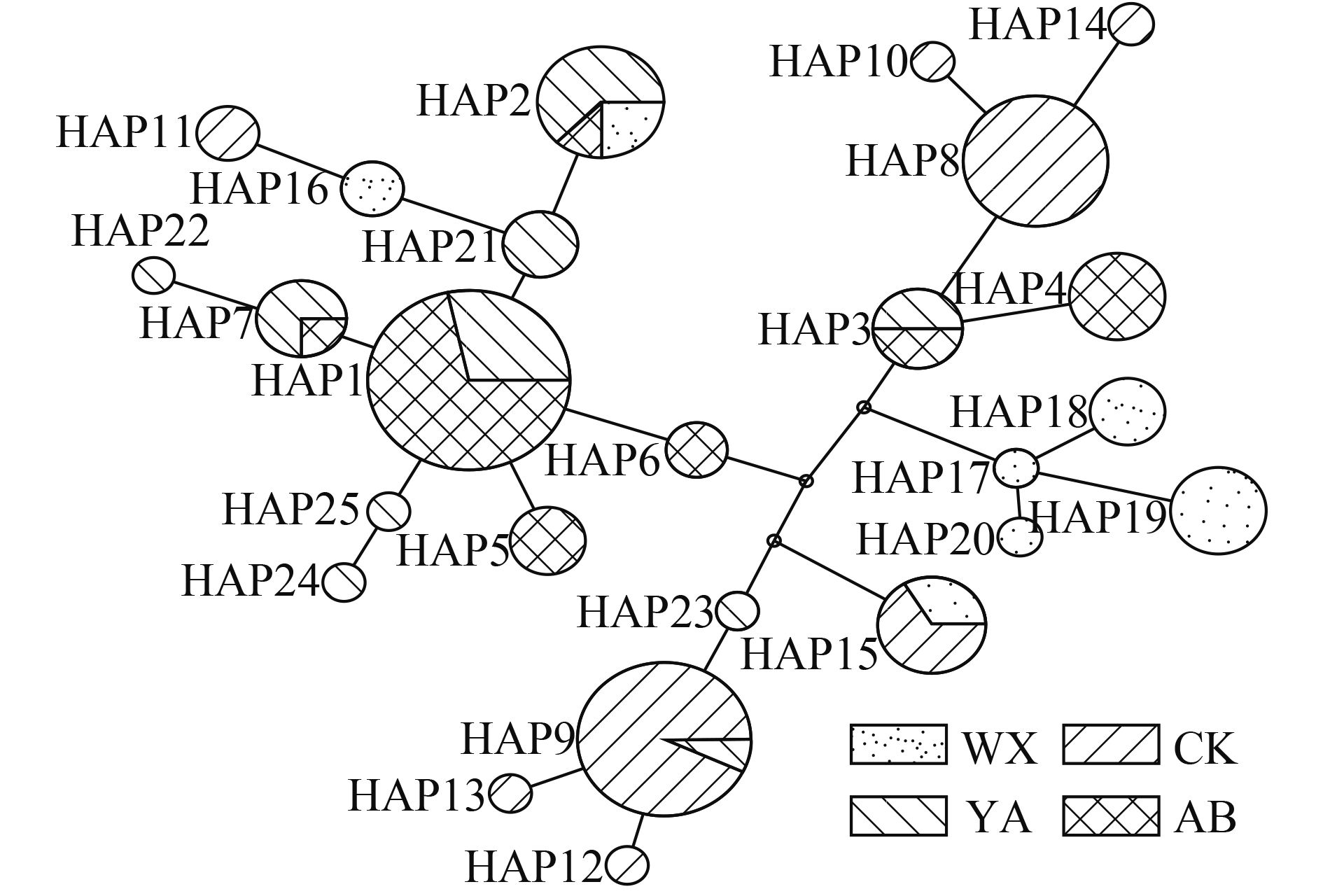
104个样本中共检测到25个单倍型, 其中Hap1、Hap8、Hap9为优势单倍型, 分别占个体数的20.18%、10.57%和14.42%; 单倍型Hap1、Hap2、Hap3、Hap7、Hap9和Hap15被2—3个群体共享, 其他19个单倍型为各群体特有单倍型, 其中, Hap2是分布最广泛的单倍型, 除了城口群体, 其他3个群体都有分布(图 1)。在4个群体中, 巫溪和雅安群体单倍型比例分别为43.75%和41.67%, 表现了较为丰富的单倍型数。而阿坝群体单倍型比例为24.14%, 表现了较为贫瘠的单倍型数。
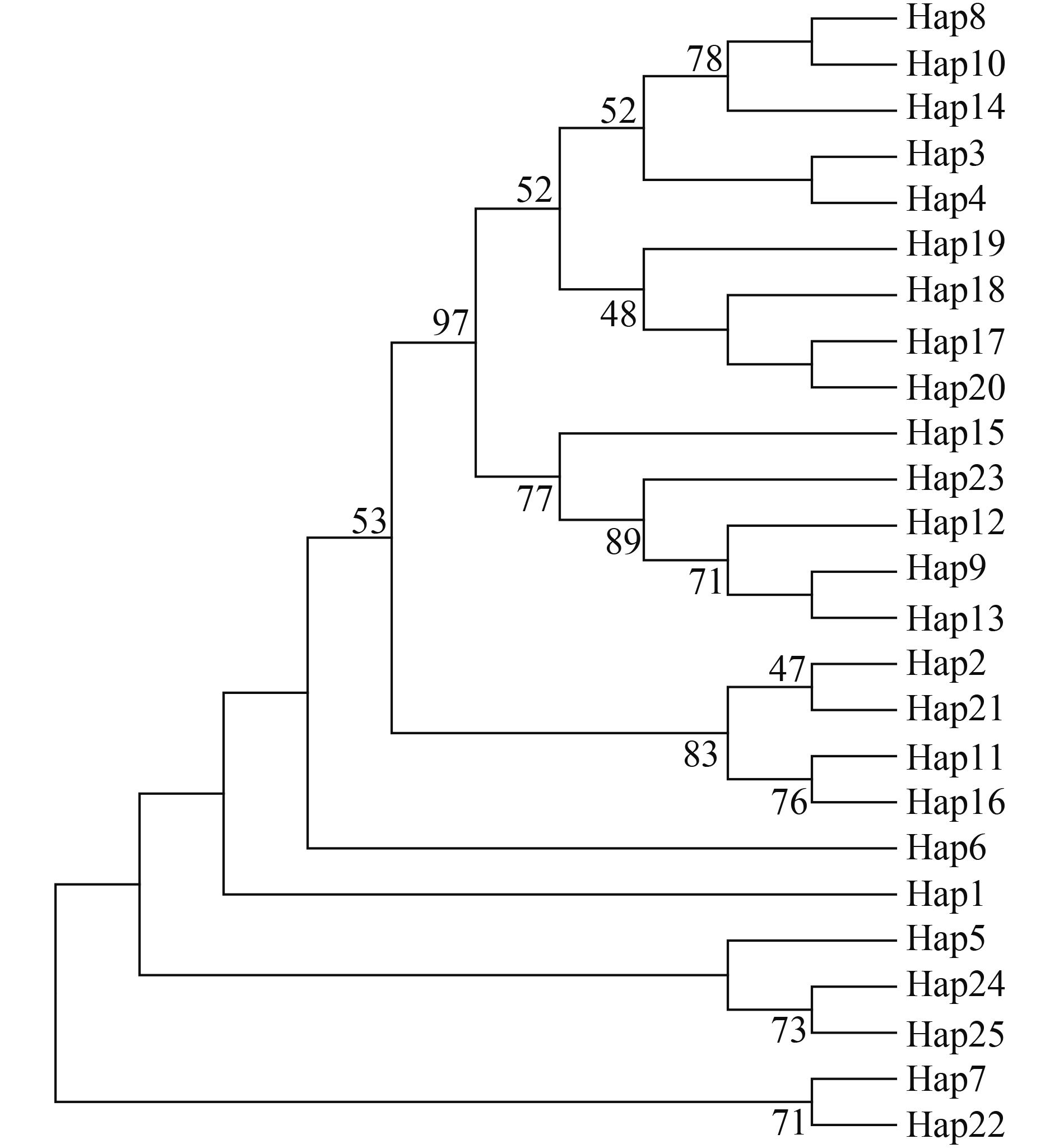
基于齐口裂腹鱼单倍型构建NJ分子系统树(图 1)可知, 25个单倍型被分成2个主支。其中一个主支包含14个单倍型, 有66个个体, 为优势分支; 另一个主支包含11个单倍型, 有48个个体。分子系统树表明, 尽管不同群体的单倍型没有严密的表现出与其地理位置分布的关系, 但是雅安和阿坝州群体之间, 以及巫溪和城口群体之间表现出了较近的亲缘关系。基于25个单倍型构建的MP分子系统树(图 2)与MJ网络图(图 3)显示的结果相吻合。
2.3 群体遗传多样性和遗传分化
巫溪和雅安群体单倍型多样性明显高于城口和阿坝群体, 其中阿坝群体的值最低(0.704±0.079); 从核苷酸多样性和平均核苷酸差异数来看, 巫溪群体的值最高(0.012±0.001; 8.033), 阿坝群体的值最低(0.007±0.001; 4.621)。从以上结果可以看出(表 2), 在4个群体中, 巫溪群体遗传多样性最高, 阿坝群体遗传多样性最低。
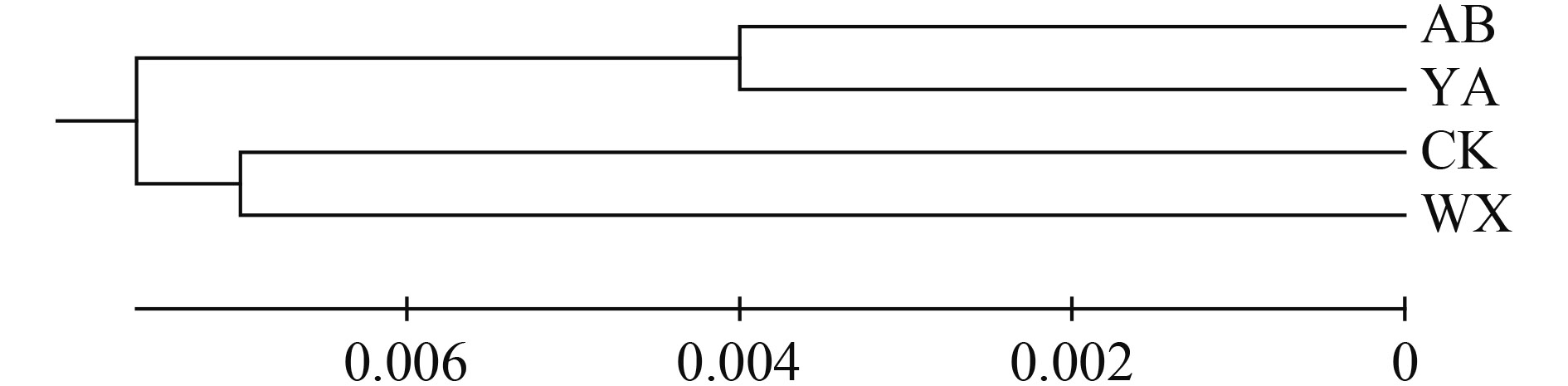
表 2 不同群体齐口裂腹鱼遗传多样性参数Table 2. Genetic diversity index of four S. prenanti populations群体 Population 多态性位点 S 单倍型数 H 单倍型多样性 Hd 核苷酸多样性 π 平均核苷酸差异数 K 重庆巫溪WX 23 7 0.867±0.057 0.0117±0.0013 8.033 重庆城口CK 29 8 0.743±0.050 0.0099±0.0010 6.824 四川雅安YA 22 10 0.884±0.038 0.0075±0.0014 5.152 四川阿坝AB 17 7 0.704±0.079 0.0067±0.0012 4.621 总体Total 43 25 0.915±0.014 0.0123±0.0004 8.492 从齐口裂腹鱼4个地理群体的个体间遗传距离来看(表 3), 群体内遗传距离大小次序为: 巫溪(0.012)>城口(0.010)>雅安(0.008)>阿坝州(0.007), 群体间的遗传距离在0.008—0.017。其中, 雅安、阿坝群体与城口群体间遗传距离较大, 雅安与阿坝群体之间遗传距离最小。这说明, 雅安群体与城口群体亲缘关系较远, 雅安群体与阿坝群体亲缘关系较近。基于群体间K2P遗传距离构建UPGMA树, 结果显示, 阿坝群体和雅安群体聚类为一支(四川组群), 巫溪群体和城口群体聚为另外一支(重庆组群)(图 4)。
表 3 齐口裂腹鱼群体内遗传距离(对角线, 粗体)及群体间遗传距离(对角线下)和遗传分化系数(Φst)(对角线上)Table 3. K2P genetic distances within population (diagonal, bold), and pairwise genetic distance (below diagonal) and fixation index (Φst) (above diagonal) among populations of S. prenanti populations重庆巫溪WX 重庆城口CK 四川雅安YA 四川阿坝AB 重庆巫溪WX 0.012 0.228** 0.299** 0.332** 重庆城口CK 0.014 0.010 0.458** 0.468** 四川雅安YA 0.014 0.017 0.008 0.054 四川阿坝AB 0.014 0.016 0.008 0.007 注: **表示经Bonferonni校正后在α=0.01水平显著 Note: ** indicates significant at α=0.01 after Bonferonni correction 齐口裂腹鱼群体间的遗传分化指数如表 3(对角线上)所示。4个群体遗传分化指数在0.054— 0.468。根据Wright关于群体遗传分化程度的理论[32], 雅安与阿坝群体之间的遗传分化指数最小(Φst=0.054, P>0.05), 其值在0.05—0.15, 属于低度分化, 表明雅安和阿坝群体间遗传分化程度较低。巫溪与城口群体之间的遗传分化指数(Φst=0.228, P=0)在0.15—0.25, 属于中度分化。城口群体与四川组群(雅安、阿坝群体)之间, 以及巫溪与四川组群(雅安、阿坝群体)之间遗传分化指数都大于0.25, 属于高度分化。其中, 城口和阿坝群体之间遗传分化指数最高(Φst=0.468, P=0)。
在AMOVA分析中(表 4), 将4个群体归为一个组群分析表明, 长江上游齐口裂腹鱼群体内的变异组分(64.23%)大于群体间的变异组分(35.77%), 各群体间存在着显著遗传分化(Fst=0.357, P=0)。基于遗传分化分析结果, 将4个群体分成四川和重庆2个组群, 结果表明, 31.50%的变异组分来自2个组群之间, 两组群间存在显著的遗传分化(Fct=0.315, P=0), 10.69%的变异组分来自组群内群体间(Fsc=0.156, P=0), 而各个群体内个体间的变异组分达到57.81% (Fst=0.422, P=0), 由此说明, 长江上游齐口裂腹鱼的变异主要来自群体内个体间, 不同组群间及组群内群体间存在显著遗传分化。
表 4 齐口裂腹鱼群体Cyt b序列变异的分子方差分析(AMOVA)Table 4. Analysis of molecular variances (AMOVA) of Cyt b sequences of S. prenanti populations变异来源 Source of variation 自由度 df 平方和 Sum of squares 方差组分 Variance components 百分率 Percentage of variation (%) 固定指数 Fixation indices P 一个组群One group 群体间Among populations 3 137.169 1.683 35.77 Fst=0.357** 0 群体内Within populations 100 302.119 3.021 64.23 两个组群Two groups 组群间Among groups 1 104.190 1.646 31.50 Fct=0.315** 0 组群内群体间Among populations within groups 2 32.979 0.559 10.69 Fsc=0.156** 0 群体内Within populations 100 302.119 3.021 57.81 Fst=0.422** 0 注: **表示在α=0.01水平显著 Note: ** indicates significant at α=0.01 对4个群体间基因流Nm分析可知(表 5), 巫溪群体与城口群体, 雅安群体与阿坝群体之间的Nm值均大于1, 说明重庆组群以及四川组群内都有基因交流。其中, 雅安群体与阿坝群体间Nm值高达8.791, 表明两群体间存在较为频繁的基因交流。而城口群体与四川组群(雅安、阿坝群体)之间的Nm值均小于1, 巫溪群体与四川组群(雅安、阿坝群体)之间的Nm值略大于1, 表明四川组群和重庆组群之间基因交流受阻。
表 5 齐口裂腹鱼群体间的基因流Nm(左下角)Table 5. Gene flow value (Nm, below diagonal) between every two populations of S. prenanti重庆巫溪WX 重庆城口CK 四川雅安YA 四川阿坝AB 重庆巫溪WX 重庆城口CK 1.696 四川雅安YA 1.174 0.592 四川阿坝AB 1.007 0.567 8.791 2.4 群体历史动态分析
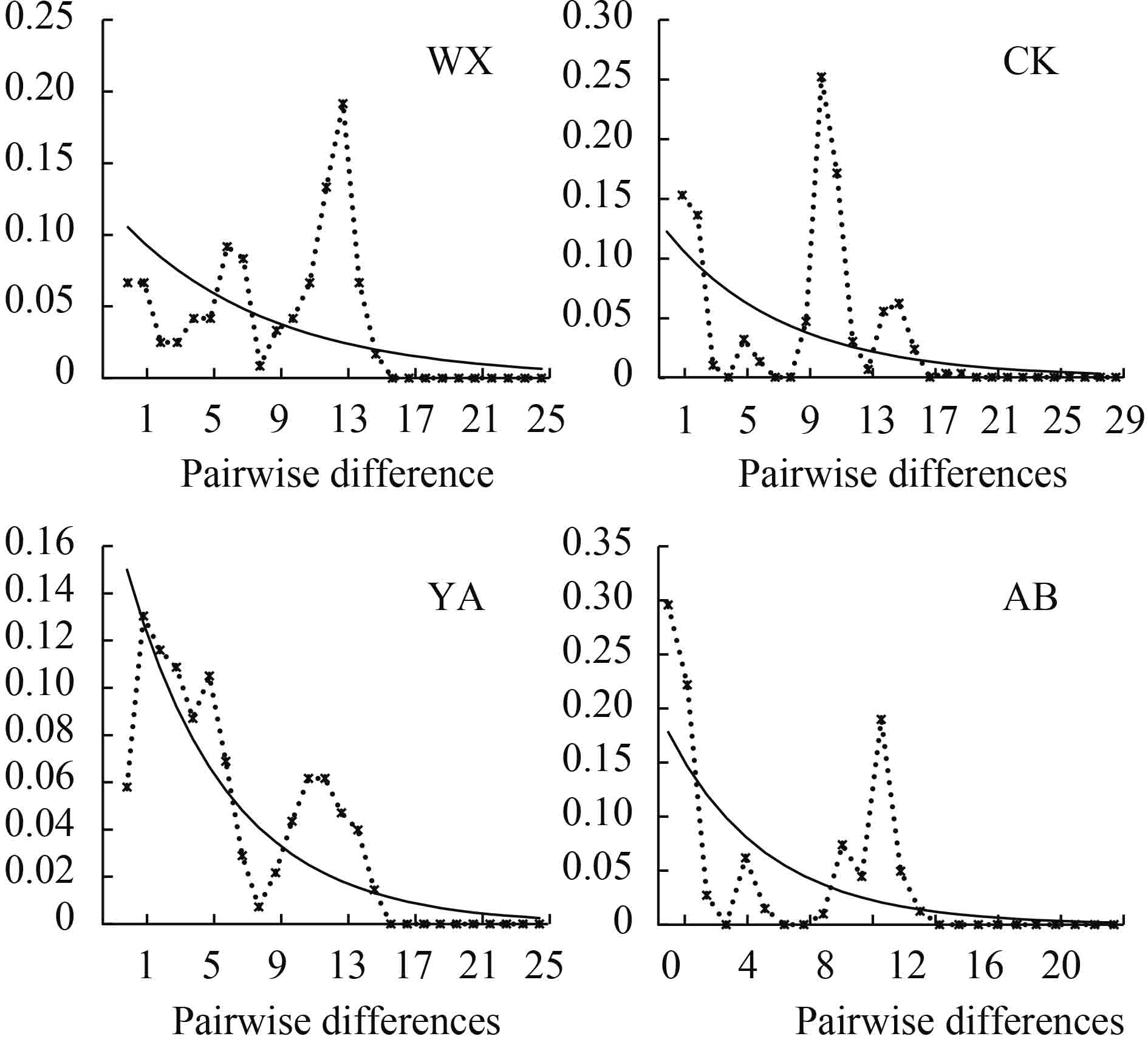
对所有个体进行中性检验(表 6)和歧点分布分析, 结果显示4个群体的Fu’s Fs检验及Tajima’s D检验结果都不显著(P>0.05), 且其歧点分布图均呈明显的多峰形(图 5), 表明4个齐口裂腹鱼野生群体在历史进化的过程中未经历过种群扩张。
表 6 齐口裂腹鱼群体的Fu’SFs和Tajima’s D中性检验Table 6. Fu’SFs and Tajima’s D of S. prenanti populations群体 Population 样本数 Sample size Fu’SFs P value Tajima’s D P value Fs P D P 重庆巫溪WX 16 2.548 0.869 0.646 0.764 重庆城口CK 35 4.203 0.929 –0.109 0.508 四川雅安YA 24 –0.083 0.509 –0.462 0.347 四川阿坝AB 29 2.461 0.873 0.232 0.633 总体Total 104 –0.898 0.452 0.095 0.617 3. 讨论
3.1 Cyt b序列变异特征
本研究对长江上游4个野生群体齐口裂腹鱼Cyt b序列分析表明, A+T达到55.20%, 明显量高于G+C含量, 这与晁燕等[33]在齐口裂腹鱼中发现的54.9% A+T含量相符。这可能与生物的进化地位有关, 即线粒体DNA的进化地位随着A+T含量的增加而增高[34]。同时, 本研究低G(16.90%)含量与Qi等[20]和谢佳燕等[35]对长江干流及青藏高原的齐口裂腹鱼的研究结果一致, 表明Cyt b有很强的碱基组成偏向性。
研究发现长江上游104个齐口裂腹鱼Cyt b序列有68个变异位点, 共检测到单倍型数为25个。单倍型数占全部个体数的比例为24.04%, 高于孟玮等[36]对塔里木裂腹鱼的研究结果(10.53%), 低于Qi等[20]对长江干流齐口裂腹鱼的研究结果(53.85%)。与谢佳燕等[35]对青藏高原齐口裂腹鱼的研究结果相似(29.63%)。25个单倍型中Hap2分布最广泛, 存在于除重庆城口以外的其他3个群体中。长江上游4个野生齐口裂腹鱼群体独享19个单倍型, 远高于晁燕等[33]的研究结果(2个), 说明长江上游四川、重庆江段干流及支流齐口裂腹鱼群体单倍型较青海班玛县玛柯河的丰富, 这些群体更能够有代表性的说明齐口裂腹鱼群体的遗传多样性。
3.2 群体遗传多样性
单倍型多样性指数(Hd)和核苷酸多样性指数(π)是评估遗传多样性的两个重要参数。本研究中的齐口裂腹鱼群体多样性指数(Hd=0.704—0.884, π=0.007—0.012)高于青海玛柯河和四川丹巴的齐口裂腹鱼群体(Hd=0.56—0.83, π=0.0007— 0.0032)[36], 低于木里河、大渡河和青衣江群体(Hd= 0.93—0.99, π=0.014—0.02)[9]; 与其他裂腹鱼亚科鱼类相比, 高于塔里木裂腹鱼(Hd=0.19—0.74, π= 0.0004—0.0029)[34], 和黄河和托索湖的黄河裸裂尻(Hd=0.67—0.70, π=0.0012—0.0026)[37], 略低于柴达木内流水系和4个黄河支流的黄河裸裂尻(Hd= 0.79—0.97, π=0.0027—0.0099)[38]。综上可知, 齐口裂腹鱼遗传多样性处于中等水平。在4个群体中, 巫溪和雅安群体的Hd和π值较高, 巫溪群体群体内遗传距离最高, 遗传变异最丰富, 遗传多样性最高, 而阿坝群体遗传变异最小, 多样性最低。重庆巫溪群体遗传多样性相对较高, 一方面可能与重庆巫溪有着较为适宜的水文条件和优越的生存环境有关; 另一方面可能是巫溪地势险峻, 人为活动对齐口裂腹鱼野生种质资源的影响较小。四川阿坝群体遗传多样性较低, 这可能与近年来自然环境的恶化及以及栖息水域受到污染、繁育环境发生改变、过度捕捞等人为活动的影响有关。
利用中性检验和核苷酸歧点分布研究物种的种群历史, 若Fu’s Fs的检测呈显著负值且歧点分布分析图谱呈泊松状分布的单峰, 说明种群趋于发生了瓶颈效应或者种群扩张[39]。本研究Fu’s Fs和Tajima’s D均不显著(P>0.05), 且歧点分布图均呈明显的多峰形, 表明长江上游4个野生齐口裂腹鱼群体在历史进化的过程中相对较为稳定, 未经历过种群扩张。
3.3 群体遗传结构
研究群体遗传结构不仅可以用于评价物种群体的变异水平和不同地理群体之间的关系, 还可以确定群体中的进化显著单元和管理单元, 以及制定资源的保护和管理策略[40]。在本研究中, 除雅安与阿坝群体其余群体间遗传分化均达极显著水平, 但群体间最大遗传距离(0.017)仍小于亚种的界限(0.02), 其均未达到亚种分化[41,42]。根据Wright[32]关于对群体遗传分化和基因交流程度的理论, 4个野生群体中, 城口和雅安群体(Φst =0.458, P=0, Nm=0.592)、城口和阿坝群体(Φst=0.468, P=0, Nm= 0.567)之间基因交流受阻, 群体间遗传分化达极显著水平, 这可能与分布流域较远和地理隔离等因素有关。阿坝和雅安群体(Φst=0.054, P>0.05Nm=8.791)基因交流频繁, 群体间遗传分化不显著。同时, 阿坝和雅安群体之间共享多个单倍型, 有较小的遗传距离, 可能是因为脚木足河是大渡河的支流, 地理分布特点决定了两群体间可频繁的进行基因交流。
本研究对齐口裂腹鱼主要分布区域, 包括长江上游四川、重庆江段干流及支流齐口裂腹鱼种群进行了多样性分析, 4个群体遗传多样性整体不高, 应该加强齐口裂腹鱼各群体的资源保护力度。由于四川组群与重庆组群之间存在显著的遗传分化, 建议将四川群体、重庆群体分别作为2个进化显著单元(Evolutionarily Significant Unit, ESU)进行保护。本研究初步反映了齐口裂腹鱼的遗传背景、遗传结构及遗传变异水平, 后续可以通过利用多种分子标记技术如SSR、SNP等联合分析, 更全面地为齐口裂腹鱼资源的保护和合理开发提供理论依据。
-
表 1 齐口裂腹鱼样本采集信息
Table 1 Sampling information of S. prenanti
群体 Population 河流 River 经纬度 Longitude and Latitude 采样时间 Time 样本数 Sample size 重庆巫溪WX 大宁河 +109.31, +31.35 2014.5— 2015.5 16 重庆城口CK 任河 +108.15, +31.37 2014.7—9 35 四川雅安YA 大渡河 +101.55, +28.51 2015.4 24 四川阿坝州AB 脚木足河 +102.22, +31.89 2014.3—4 29 表 2 不同群体齐口裂腹鱼遗传多样性参数
Table 2 Genetic diversity index of four S. prenanti populations
群体 Population 多态性位点 S 单倍型数 H 单倍型多样性 Hd 核苷酸多样性 π 平均核苷酸差异数 K 重庆巫溪WX 23 7 0.867±0.057 0.0117±0.0013 8.033 重庆城口CK 29 8 0.743±0.050 0.0099±0.0010 6.824 四川雅安YA 22 10 0.884±0.038 0.0075±0.0014 5.152 四川阿坝AB 17 7 0.704±0.079 0.0067±0.0012 4.621 总体Total 43 25 0.915±0.014 0.0123±0.0004 8.492 表 3 齐口裂腹鱼群体内遗传距离(对角线, 粗体)及群体间遗传距离(对角线下)和遗传分化系数(Φst)(对角线上)
Table 3 K2P genetic distances within population (diagonal, bold), and pairwise genetic distance (below diagonal) and fixation index (Φst) (above diagonal) among populations of S. prenanti populations
重庆巫溪WX 重庆城口CK 四川雅安YA 四川阿坝AB 重庆巫溪WX 0.012 0.228** 0.299** 0.332** 重庆城口CK 0.014 0.010 0.458** 0.468** 四川雅安YA 0.014 0.017 0.008 0.054 四川阿坝AB 0.014 0.016 0.008 0.007 注: **表示经Bonferonni校正后在α=0.01水平显著 Note: ** indicates significant at α=0.01 after Bonferonni correction 表 4 齐口裂腹鱼群体Cyt b序列变异的分子方差分析(AMOVA)
Table 4 Analysis of molecular variances (AMOVA) of Cyt b sequences of S. prenanti populations
变异来源 Source of variation 自由度 df 平方和 Sum of squares 方差组分 Variance components 百分率 Percentage of variation (%) 固定指数 Fixation indices P 一个组群One group 群体间Among populations 3 137.169 1.683 35.77 Fst=0.357** 0 群体内Within populations 100 302.119 3.021 64.23 两个组群Two groups 组群间Among groups 1 104.190 1.646 31.50 Fct=0.315** 0 组群内群体间Among populations within groups 2 32.979 0.559 10.69 Fsc=0.156** 0 群体内Within populations 100 302.119 3.021 57.81 Fst=0.422** 0 注: **表示在α=0.01水平显著 Note: ** indicates significant at α=0.01 表 5 齐口裂腹鱼群体间的基因流Nm(左下角)
Table 5 Gene flow value (Nm, below diagonal) between every two populations of S. prenanti
重庆巫溪WX 重庆城口CK 四川雅安YA 四川阿坝AB 重庆巫溪WX 重庆城口CK 1.696 四川雅安YA 1.174 0.592 四川阿坝AB 1.007 0.567 8.791 表 6 齐口裂腹鱼群体的Fu’SFs和Tajima’s D中性检验
Table 6 Fu’SFs and Tajima’s D of S. prenanti populations
群体 Population 样本数 Sample size Fu’SFs P value Tajima’s D P value Fs P D P 重庆巫溪WX 16 2.548 0.869 0.646 0.764 重庆城口CK 35 4.203 0.929 –0.109 0.508 四川雅安YA 24 –0.083 0.509 –0.462 0.347 四川阿坝AB 29 2.461 0.873 0.232 0.633 总体Total 104 –0.898 0.452 0.095 0.617 -
[1] 武云飞, 吴翠珍. 青藏高原鱼类. 成都: 四川科学技术出版社. 1992, 361—365 Wu Y F, Wu C Z. The Qinghai-Tibet Plateau Fish [M]. Chengdu: Sichuan Science and Technology Press. 1992, 361—365
武云飞, 吴翠珍. 青藏高原鱼类. 成都: 四川科学技术出版社. 1992, 361—365[2] 陈毅峰, 曹文宣. 裂腹鱼亚科. 乐佩琦, 单乡红, 林人端, 等. 中国动物志•硬骨鱼纲•鲤形目(下). 北京: 科学出版社. 2000, 273—390 Chen Y F, Cao W X. Schizothoracinae. In: Le P Q, Shan X H, Lin R D, et al. Chinese Animal: Osteichthyes Carp Shape Mesh Volume [M]. Beijing: Science and Technology Press. 2000, 273—390
陈毅峰, 曹文宣. 裂腹鱼亚科. 乐佩琦, 单乡红, 林人端, 等. 中国动物志•硬骨鱼纲•鲤形目(下). 北京: 科学出版社. 2000, 273—390[3] 刘绍平, 段辛斌, 陈大庆, 等. 长江中游渔业资源现状研究. 水生生物学报, 2005, 29(6): 708—711 http://ssswxb.ihb.ac.cn/CN/abstract/abstract3162.shtml Liu S P, Duan X B, Chen D Q, et al. Studies on status of fishery resources in the middle reach of the Yangtze River [J]. Acta Hydrobiologica Sinica, 2005, 29(6): 708—711
刘绍平, 段辛斌, 陈大庆, 等. 长江中游渔业资源现状研究. 水生生物学报, 2005, 29(6): 708—711 http://ssswxb.ihb.ac.cn/CN/abstract/abstract3162.shtml[4] 若木, 王鸿泰, 殷启云, 等. 齐口裂腹鱼人工繁殖的研究. 淡水渔业, 2001, 31(6): 3—5 http://www.cnki.com.cn/Article/CJFDTOTAL-DSYY201503009.htm Ruo M, W H T, Y Q Y, et al. Artificial breeding research of Schizothorax prenanti [J]. Freshwater Fisheries, 2001, 31(6): 3—5
若木, 王鸿泰, 殷启云, 等. 齐口裂腹鱼人工繁殖的研究. 淡水渔业, 2001, 31(6): 3—5 http://www.cnki.com.cn/Article/CJFDTOTAL-DSYY201503009.htm[5] Wang T, Zhou C W, Yuan D Y, et al. Schizothorax pre-nanti corticotropin-releasing hormone (CRH): molecular cloning, tissue expression, and the function of feeding regulation [J]. Fish Physiology & Biochemistry, 2014, 40(5): 1407—1415
[6] Lin F J, Zhou C W, Chen H, et al. Molecular characteri-zation, tissue distribution and feeding related changes of NUCB2A/nesfatin-1 in Ya-fish (Schizothorax prenanti) [J]. Gene, 2014, 536(2): 238—246 doi: 10.1016/j.gene.2013.12.031
[7] Zhang L, Wu Y L, Wang L, et al. Effects of Oxidized Konjac glucomannan (OKGM) on growth and immune function of Schizothorax prenanti [J]. Fish & Shellfish Immunology, 2013, 35(4): 1105—1110
[8] Song J, Song Z B, Yue B S, et al. Assessing genetic diversity of wild populations of Prenant’s schizothoracin, Schizothorax prenanti, using AFLP markers [J]. Environmental Biology of Fishes, 2006, 77(1): 79—86 doi: 10.1007/s10641-006-9056-x
[9] Song Z B, Song J, Yue B S. Population genetic diversity of Prenant’s schizothoracin Schizothorax prenanti, inferred from the mitochondrial DNA control region [J]. Environmental Biology of Fishes, 2008, 81(3): 247—252 doi: 10.1007/s10641-007-9197-6
[10] Liang J J, Liu Y, Zhang X F. An observation of the loss of genetic variability in Schizothorax prenanti, Schizothorax prenanti, inhabiting a plateau lake [J]. Biochemi-cal Systematics and Ecology, 2011, 39(4—6): 361—370 doi: 10.1016/j.bse.2011.05.005
[11] Meland S, Johansen S, Johansen T, et al. Rapid disappearance of one parental mitochondrial genotype alfter isogamous mating in the myxomycete Physarum polycephalum [J]. Current Genetics, 1991, 19(1): 55—60 doi: 10.1007/BF00362088
[12] 赵凯. 鱼类线粒体DNA 及其在分子系统学中的应用. 青海大学学报: 自然科学版, 2006, 24(4): 49—53 http://www.cnki.com.cn/Article/CJFDTOTAL-QHXZ200602013.htm Zhao K. Mitochondrial DNA (mtDNA) of fish and appli-cation in molecular phylogenetic [J]. Journal of Qinghai University (Nature Science), 2006, 24(4): 49—53
赵凯. 鱼类线粒体DNA 及其在分子系统学中的应用. 青海大学学报: 自然科学版, 2006, 24(4): 49—53 http://www.cnki.com.cn/Article/CJFDTOTAL-QHXZ200602013.htm[13] Lu X, Wang H, Liu B, et al. An effective method for pa-rentage determination of the clam (Meretrix meretrix) based on SSR and COI markers [J]. Aquaculture, 2011, 318(1—2): 223—228 doi: 10.1016/j.aquaculture.2011.05.017
[14] Mabuchi K, Miya M, Senou H, et al. Complete mitochondrial DNA sequence of the Lake Biwa wild strain of common carp (Cyprinus carpio L.): further evidence for an ancient origin [J]. Aquaculture, 2006, 257(1—4): 67—77
[15] 肖武汉, 张亚平. 鱼类线粒体DNA的遗传与进化. 水生生物学报, 2000, 24(4): 384—391 http://ssswxb.ihb.ac.cn/CN/abstract/abstract611.shtml Xiao W H, Zhang Y P. Genetics and evolution of mitochondrial DNA in fish [J]. Acta Hydrobiologica Sinica, 2000, 24(4): 384—391
肖武汉, 张亚平. 鱼类线粒体DNA的遗传与进化. 水生生物学报, 2000, 24(4): 384—391 http://ssswxb.ihb.ac.cn/CN/abstract/abstract611.shtml[16] 郭新红, 刘少军, 刘巧, 等. 鱼类线粒体DNA研究新进展. 遗传学报, 2004, 31(9): 983—1000 http://www.cnki.com.cn/Article/CJFDTOTAL-YCXB200409016.htm Guo X H, Liu S J, Liu Q, et al. New progresses on mitochondrial DNA in fish [J]. Acta Genetica Sinica, 2004, 31(9): 983—1000
郭新红, 刘少军, 刘巧, 等. 鱼类线粒体DNA研究新进展. 遗传学报, 2004, 31(9): 983—1000 http://www.cnki.com.cn/Article/CJFDTOTAL-YCXB200409016.htm[17] 胡静, 侯新远, 尹绍武, 等. 基于mtDNA COI和Cyt b基因序列对中国南海不同海域波纹唇鱼群体遗传多样性的研究. 水生生物学报, 2014, 38(6): 1008—1016 doi: 10.7541/2014.149 Hu J, Hou X Y, Yin S W, et al. Genetic diversity and divergence of Cheilinus undulates of different geographic populations of the south China sea revealed by COI and CYTB gene analyses [J]. Acta Hydrobiologica Sinica, 2014, 38(6): 1008—1016
胡静, 侯新远, 尹绍武, 等. 基于mtDNA COI和Cyt b基因序列对中国南海不同海域波纹唇鱼群体遗传多样性的研究. 水生生物学报, 2014, 38(6): 1008—1016 doi: 10.7541/2014.149[18] Mcveigh H P, Davidson W S. A salmonid phylogeny inferred from mitochondrial cytochrome b gene sequences [J]. Journal of Fish Biology, 2006, 39(sA): 277—282
[19] Durand J D, Tsigenopoulos C S, Unlu E, et al. Phylogeny and biogeography of the family cyprinidae in the middle east inferred from cytochrome b DNA- evolutio-nary significance of this region [J]. Molecular Phylogene-tics & Evolution, 2002, 22(1): 91—100
[20] Qi D L, Li T P, Zhao X Q, et al. Mitochondrial cytochrome b sequence variation and phylogenetics of the highly specialized Schizothoracine fishes (Teleostei: Cyprinidae) in the Qinghai-Tibet Plateau [J]. Biochemical Genetics, 2006, 744(5): 270—285
[21] Chen C N, Huang Y Y, Li H, et al. The complete mitochondrial genome of Schizothorax prenanti (Tchang) (Teleostei, Cyprinidae, Schizothoracinae) [J]. Mitochondrial DNA, 2016, 27(1): 253—254 doi: 10.3109/19401736.2014.883612
[22] Hall T A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT [J]. Nucleic Acids Symposium Series, 1998, 41: 95—98
[23] Thompson J D, Gibson T J, Plewniak F, et al. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools [J]. Nucleic Acids Research, 1997, 25(24): 4876—4882 doi: 10.1093/nar/25.24.4876
[24] Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data [J]. Bioinformatics, 2009, 25(11): 1451—1452(2) doi: 10.1093/bioinformatics/btp187
[25] Excoffier L, Lischer H E L. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows [J]. Molecular Ecology Resources, 2010, 10(3): 564—567 doi: 10.1111/men.2010.10.issue-3
[26] Bandelt H J, Forster P, Röhl A A. Median-joining network for inferring intraspecific phylogenies [J]. Molecular Biology & Evolution, 1999, 16(1): 37—48
[27] Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods [J]. Molecular Biology and Evolution, 2011, 28(10): 2731—2739 doi: 10.1093/molbev/msr121
[28] Hickerson M J, Meyer C P. Testing comparative phylogeographic models of marine vicariance and dispersal using a hierarchical Bayesian approach [J]. BMC Evolutio-nary Biology, 2007, 8(3): 322—340
[29] Fu Y X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection [J]. Genetics, 1997, 147(2): 915—925
[30] Aris-Brosou S, Excoffier L. The impact of population expansion and mutation rate heterogeneity on DNA sequence polymorphism [J]. Molecular Biology and Evolution, 1996, 13(3): 494—504 doi: 10.1093/oxfordjournals.molbev.a025610
[31] Rice W R. Analyzing tables of statistical tests [J]. Evolution, 1989, 43(1): 223—225 doi: 10.2307/2409177
[32] Wright S. Evolution in Mendelian populations [J]. Bulletin of Mathematical Biology, 1990, 52(1—2): 241—295 doi: 10.1007/BF02459575
[33] 晁燕, 杨成, 申志新, 等. 齐口裂腹鱼基于Cyt b基因序列多态性研究. 水产科学, 2010, 29(7): 421—424 Chao Y, Yang C, Shen Z X, et al. Genetic Diversity of Schizothorax prenanti inferred from mitochondrial cytochrome b gene sequences [J]. Fisheries Science, 2010, 29(7): 421—424
晁燕, 杨成, 申志新, 等. 齐口裂腹鱼基于Cyt b基因序列多态性研究. 水产科学, 2010, 29(7): 421—424[34] Folmer O, Black M, Hoeh W, et al. DNA primer for amplification of mitochondrial cytochrome coxidase subunit I from diverse metazoan invertebrates [J]. Molecular Ma-rine Biotechnology, 1994, 3(5): 294—299
[35] 谢佳燕. 齐口裂腹鱼线粒体DNA控制区结构分析. 动物学杂志, 2011, 46(2): 97—101 http://www.cnki.com.cn/Article/CJFDTOTAL-BIRD201102016.htm Xie J Y. Structure analysis of mtDNA control region in Schizothorax prenanti [J]. Chinese Journal of Zoology, 2011, 46(2): 97—101
谢佳燕. 齐口裂腹鱼线粒体DNA控制区结构分析. 动物学杂志, 2011, 46(2): 97—101 http://www.cnki.com.cn/Article/CJFDTOTAL-BIRD201102016.htm[36] 孟玮, 郭焱, 海萨, 等. 塔里木裂腹鱼群体遗传结构及遗传多样性分析. 水生生物学报, 2012, 36(5): 851—857 http://ssswxb.ihb.ac.cn/CN/abstract/abstract2159.shtml Meng W, Guo Y, Hai S, et al. Genetic structure and diversity of Schizothorax bidduphi populations [J]. Acta Hydrobiologica Sinica, 2012, 36(5): 851—857
孟玮, 郭焱, 海萨, 等. 塔里木裂腹鱼群体遗传结构及遗传多样性分析. 水生生物学报, 2012, 36(5): 851—857 http://ssswxb.ihb.ac.cn/CN/abstract/abstract2159.shtml[37] 赵凯, 杨公社, 李俊冰, 等. 黄河裸裂尻鱼群体遗传结构和Cyt b序列变异. 水生生物学报, 2006, 30(2): 129—133 http://ssswxb.ihb.ac.cn/CN/abstract/abstract2542.shtml Zhao K, Yang G S, Li J B, et al. Phylogenetic structure of Schizopygopsis pylzovi populations from mitochondrial B gene sequence variations [J]. Acta Hydrobiologica Sinica, 2006, 30(2): 129—133
赵凯, 杨公社, 李俊冰, 等. 黄河裸裂尻鱼群体遗传结构和Cyt b序列变异. 水生生物学报, 2006, 30(2): 129—133 http://ssswxb.ihb.ac.cn/CN/abstract/abstract2542.shtml[38] 祁得林, 晁燕, 郭松, 等. 黄河裸裂尻鱼五种群mtDNA控制区的遗传结构. 动物学报, 2008, 54(6): 972—980 http://www.cnki.com.cn/Article/CJFDTOTAL-BEAR200806005.htm Qi D L, Chao Y, Guo S, et al. Genetic structure of five Huanghe schizothoracin Schizopygopsis pylzovi populations based on mtDNA control region sequences [J]. Acta Zoologica Sinica, 2008, 54(6): 972—980
祁得林, 晁燕, 郭松, 等. 黄河裸裂尻鱼五种群mtDNA控制区的遗传结构. 动物学报, 2008, 54(6): 972—980 http://www.cnki.com.cn/Article/CJFDTOTAL-BEAR200806005.htm[39] Barbosa A M, Real R, Muñoz A R, et al. New measures for assessing model equilibrium and prediction mismatch in species distribution models [J]. Diversity and Distributions, (Diversity Distrib.) 2013, 19(10): 1333—1338 doi: 10.1111/ddi.2013.19.issue-10
[40] O’Brien S J. A role for molecular genetics in biological conservation [J]. Proceedings of the National Academy of Sciences of the United States of America, 1994, 91(13): 5748—5755 doi: 10.1073/pnas.91.13.5748
[41] Shaklee J B, Tamaru C S, Waples R S. Speciation and evolution of marine fishes studied by the electrophore-tic analysis of proteins [J]. Pacificence, 1982, 36(2): 141—157
[42] Nei, Masatoshi. Molecular Population Genetics and Evo-lution [M]. American Elsevier Publishing Company. 1975, 128—132
-
期刊类型引用(16)
1. 许保可,阿琳林,张海琛,马清花,梁健. 青海湖裸鲤TNNI1和TNNI2基因克隆及其响应盐碱胁迫的表达研究. 西北农林科技大学学报(自然科学版). 2023(10): 1-10 .  百度学术
百度学术
2. 李大命,刘洋,唐晟凯,刘燕山,蔡永久,谷先坤,殷稼雯,蒋琦辰,张彤晴. 基于Cytb基因的滆湖鲌类国家级水产种质资源保护区3种鲌鱼的遗传多样性分析. 水产科技情报. 2022(01): 1-7 .  百度学术
百度学术
3. 刘伟,曾圣,王雪,詹会祥,周路,熊畔,向燕. 基于线粒体D-loop区和Cyt b基因序列的四川裂腹鱼养殖群体遗传多样性. 西南农业学报. 2022(02): 475-480 .  百度学术
百度学术
4. 杨朝杰,高强,刘丹,聂苗苗,王发艳,李柯懋,赵宏,张存芳,祁得林. 基于mtDNA D-loop部分序列的软刺裸裂尻鱼遗传多样性分析. 四川动物. 2022(04): 398-405 .  百度学术
百度学术
5. 杨鑫,黄坤,胡靖蕊,曾丽雯,杨世勇,武佳韵. 周公河齐口裂腹鱼种群的遗传多样性和遗传结构. 水生生物学报. 2022(11): 1712-1718 .  本站查看
本站查看
6. 朱其广,唐会元,林晖,龚云,李析男,杨志. 金沙江中下游细鳞裂腹鱼的年龄生长及种群动态. 水生态学杂志. 2021(02): 56-63 .  百度学术
百度学术
7. 李大命,唐晟凯,刘燕山,谷先坤,殷稼雯,蒋琦辰,张彤晴,潘建林. 基于Cyt b基因的江苏省湖鲚种群遗传多样性和遗传结构分析. 中国农学通报. 2021(20): 144-151 .  百度学术
百度学术
8. 杨慧兰,王银肖,谭慧敏,穆淑梅,康现江,陈咏霞. 白洋淀流域宽鳍遗传多样性及种群历史动态. 上海海洋大学学报. 2021(05): 837-846 .  百度学术
百度学术
9. 孔杰,刘伟,李世凯,周洲,赵凤. 基于线粒体D-loop区序列和Cyt b基因的鲟鱼群体遗传多样性分析. 西南农业学报. 2020(01): 206-210 .  百度学术
百度学术
10. 李大命,唐晟凯,刘燕山,谷先坤,刘小维,殷稼雯,张彤晴,潘建林. 江苏省4个太湖新银鱼种群遗传多样性和遗传结构分析. 渔业科学进展. 2020(05): 52-60 .  百度学术
百度学术
11. 邵科,熊美华,阙延福,徐念,廖小林,朱滨,李伟涛. 基于线粒体Cyt b序列的雅砻江长丝裂腹鱼群体遗传结构分析. 长江流域资源与环境. 2020(12): 2686-2695 .  百度学术
百度学术
12. 孔杰,周洲,刘伟,赵凤,王金乐. 达氏鳇亲鱼群体mtDNA Cyt b基因和D-loop区的序列特征及遗传多样性分析. 江苏农业科学. 2019(03): 40-42 .  百度学术
百度学术
13. 周华兴,胡玉婷,段国庆,凌俊,江河,潘庭双. 基于线粒体细胞色素b基因序列的新安江流域温州光唇鱼群体遗传研究. 渔业科学进展. 2019(02): 43-50 .  百度学术
百度学术
14. 刘欢,刘玉芬,刘鹏,赵文阁. 胎生蜥蜴线粒体Cyt b基因和D-loop区序列多态性分析. 黑龙江畜牧兽医. 2018(14): 200-202 .  百度学术
百度学术
15. 杨月静,向梦斌,叶祥益,张争世,罗辉,叶华. 齐口裂腹鱼SNP标记与生长性状的关联分析. 中国水产科学. 2018(02): 278-285 .  百度学术
百度学术
16. 边力,王鹏飞,陈四清,李凤辉,张乐乐,刘长琳,葛建龙. 基于线粒体Cyt b基因序列的绿鳍马面鲀6个野生群体的遗传结构分析. 中国水产科学. 2018(04): 827-836 .  百度学术
百度学术
其他类型引用(13)



 下载:
下载: